MLflow Models
An MLflow Model is a standard format for packaging machine learning models that can be used in a variety of downstream tools—for example, real-time serving through a REST API or batch inference on Apache Spark. The format defines a convention that lets you save a model in different “flavors” that can be understood by different downstream tools.
Table of Contents
Storage Format
Each MLflow Model is a directory containing arbitrary files, together with an MLmodel
file in the root of the directory that can define multiple flavors that the model can be viewed
in.
The model aspect of the MLflow Model can either be a serialized object (e.g., a pickled scikit-learn model)
or a Python script (or notebook, if running in Databricks) that contains the model instance that has been defined
with the mlflow.models.set_model() API.
Flavors are the key concept that makes MLflow Models powerful: they are a convention that deployment
tools can use to understand the model, which makes it possible to write tools that work with models
from any ML library without having to integrate each tool with each library. MLflow defines
several “standard” flavors that all of its built-in deployment tools support, such as a “Python
function” flavor that describes how to run the model as a Python function. However, libraries can
also define and use other flavors. For example, MLflow’s mlflow.sklearn library allows
loading models back as a scikit-learn Pipeline object for use in code that is aware of
scikit-learn, or as a generic Python function for use in tools that just need to apply the model
(for example, the mlflow deployments tool with the option -t sagemaker for deploying models
to Amazon SageMaker).
MLmodel file
All of the flavors that a particular model supports are defined in its MLmodel file in YAML
format. For example, running python examples/sklearn_logistic_regression/train.py from MLflow repo
will create the following files under the model directory:
# Directory written by mlflow.sklearn.save_model(model, "model", input_example=...)
model/
├── MLmodel
├── model.pkl
├── conda.yaml
├── python_env.yaml
├── requirements.txt
├── input_example.json (optional, only logged when input example is provided and valid during model logging)
├── serving_input_example.json (optional, only logged when input example is provided and valid during model logging)
└── environment_variables.txt (optional, only logged when environment variables are used during model inference)
And its MLmodel file describes two flavors:
time_created: 2018-05-25T17:28:53.35
flavors:
sklearn:
sklearn_version: 0.19.1
pickled_model: model.pkl
python_function:
loader_module: mlflow.sklearn
Apart from a flavors field listing the model flavors, the MLmodel YAML format can contain the following fields:
time_created: Date and time when the model was created, in UTC ISO 8601 format.run_id: ID of the run that created the model, if the model was saved using MLflow Tracking.signature: model signature in JSON format.input_example: reference to an artifact with input example.databricks_runtime: Databricks runtime version and type, if the model was trained in a Databricks notebook or job.mlflow_version: The version of MLflow that was used to log the model.
Additional Logged Files
For environment recreation, we automatically log conda.yaml, python_env.yaml, and requirements.txt files whenever a model is logged.
These files can then be used to reinstall dependencies using conda or virtualenv with pip. Please see
How MLflow Model Records Dependencies for more details about these files.
If a model input example is provided when logging the model, two additional files input_example.json and serving_input_example.json are logged.
See Model Input Example for more details.
When logging a model, model metadata files (MLmodel, conda.yaml, python_env.yaml, requirements.txt) are copied to a subdirectory named metadata. For wheeled models, original_requirements.txt file is also copied.
Note
When a model registered in the MLflow Model Registry is downloaded, a YAML file named registered_model_meta is added to the model directory on the downloader’s side. This file contains the name and version of the model referenced in the MLflow Model Registry, and will be used for deployment and other purposes.
Attention
If you log a model within Databricks, MLflow also creates a metadata subdirectory within
the model directory. This subdirectory contains the lightweight copy of aforementioned
metadata files for internal use.
Environment variables file
MLflow records the environment variables that are used during model inference in environment_variables.txt file when logging a model.
Attention
environment_variables.txt file only contains names of the environment variables that are used during model inference,
values are not stored.
Currently MLflow only logs the environment variables whose name contains any of the following keywords:
RECORD_ENV_VAR_ALLOWLIST = {
# api key related
"API_KEY", # e.g. OPENAI_API_KEY
"API_TOKEN",
# databricks auth related
"DATABRICKS_HOST",
"DATABRICKS_USERNAME",
"DATABRICKS_PASSWORD",
"DATABRICKS_TOKEN",
"DATABRICKS_INSECURE",
"DATABRICKS_CLIENT_ID",
"DATABRICKS_CLIENT_SECRET",
"_DATABRICKS_WORKSPACE_HOST",
"_DATABRICKS_WORKSPACE_ID",
}
Example of a pyfunc model that uses environment variables:
import mlflow
import os
os.environ["TEST_API_KEY"] = "test_api_key"
class MyModel(mlflow.pyfunc.PythonModel):
def predict(self, context, model_input, params=None):
if os.environ.get("TEST_API_KEY"):
return model_input
raise Exception("API key not found")
with mlflow.start_run():
model_info = mlflow.pyfunc.log_model(
"model", python_model=MyModel(), input_example="data"
)
Environment variable TEST_API_KEY is logged in the environment_variables.txt file like below
# This file records environment variable names that are used during model inference.
# They might need to be set when creating a serving endpoint from this model.
# Note: it is not guaranteed that all environment variables listed here are required
TEST_API_KEY
Attention
Before you deploy a model to a serving endpoint, review the environment_variables.txt file to ensure all necessary environment variables for model inference are set. Note that not all environment variables listed in the file are always required for model inference. For detailed instructions on setting environment variables on a databricks serving endpoint, refer to this guidance.
Note
To disable this feature, set the environment variable MLFLOW_RECORD_ENV_VARS_IN_MODEL_LOGGING to false.
Managing Model Dependencies
An MLflow Model infers dependencies required for the model flavor and automatically logs them. However, it also allows you to define extra dependencies or custom Python code, and offer a tool to validate them in a sandbox environment. Please refer to Managing Dependencies in MLflow Models for more details.
Model Signatures And Input Examples
In MLflow, understanding the intricacies of model signatures and input examples is crucial for effective model management and deployment.
Model Signature: Defines the schema for model inputs, outputs, and additional inference parameters, promoting a standardized interface for model interaction.
Model Input Example: Provides a concrete instance of valid model input, aiding in understanding and testing model requirements. Additionally, if an input example is provided when logging a model, a model signature will be automatically inferred and stored if not explicitly provided.
Model Serving Payload Example: Provides a json payload example for querying a deployed model endpoint. If an input example is provided when logging a model, a serving paylod example is automatically generated from the input example and saved as
serving_input_example.json.
Our documentation delves into several key areas:
Supported Signature Types: We cover the different data types that are supported, such as tabular data for traditional machine learning models and tensors for deep learning models.
Signature Enforcement: Discusses how MLflow enforces schema compliance, ensuring that the provided inputs match the model’s expectations.
Logging Models with Signatures: Guides on how to incorporate signatures when logging models, enhancing clarity and reliability in model operations.
For a detailed exploration of these concepts, including examples and best practices, visit the Model Signatures and Examples Guide. If you would like to see signature enforcement in action, see the notebook tutorial on Model Signatures to learn more.
Model API
You can save and load MLflow Models in multiple ways. First, MLflow includes integrations with
several common libraries. For example, mlflow.sklearn contains
save_model, log_model,
and load_model functions for scikit-learn models. Second,
you can use the mlflow.models.Model class to create and write models. This
class has four key functions:
add_flavorto add a flavor to the model. Each flavor has a string name and a dictionary of key-value attributes, where the values can be any object that can be serialized to YAML.saveto save the model to a local directory.logto log the model as an artifact in the current run using MLflow Tracking.loadto load a model from a local directory or from an artifact in a previous run.
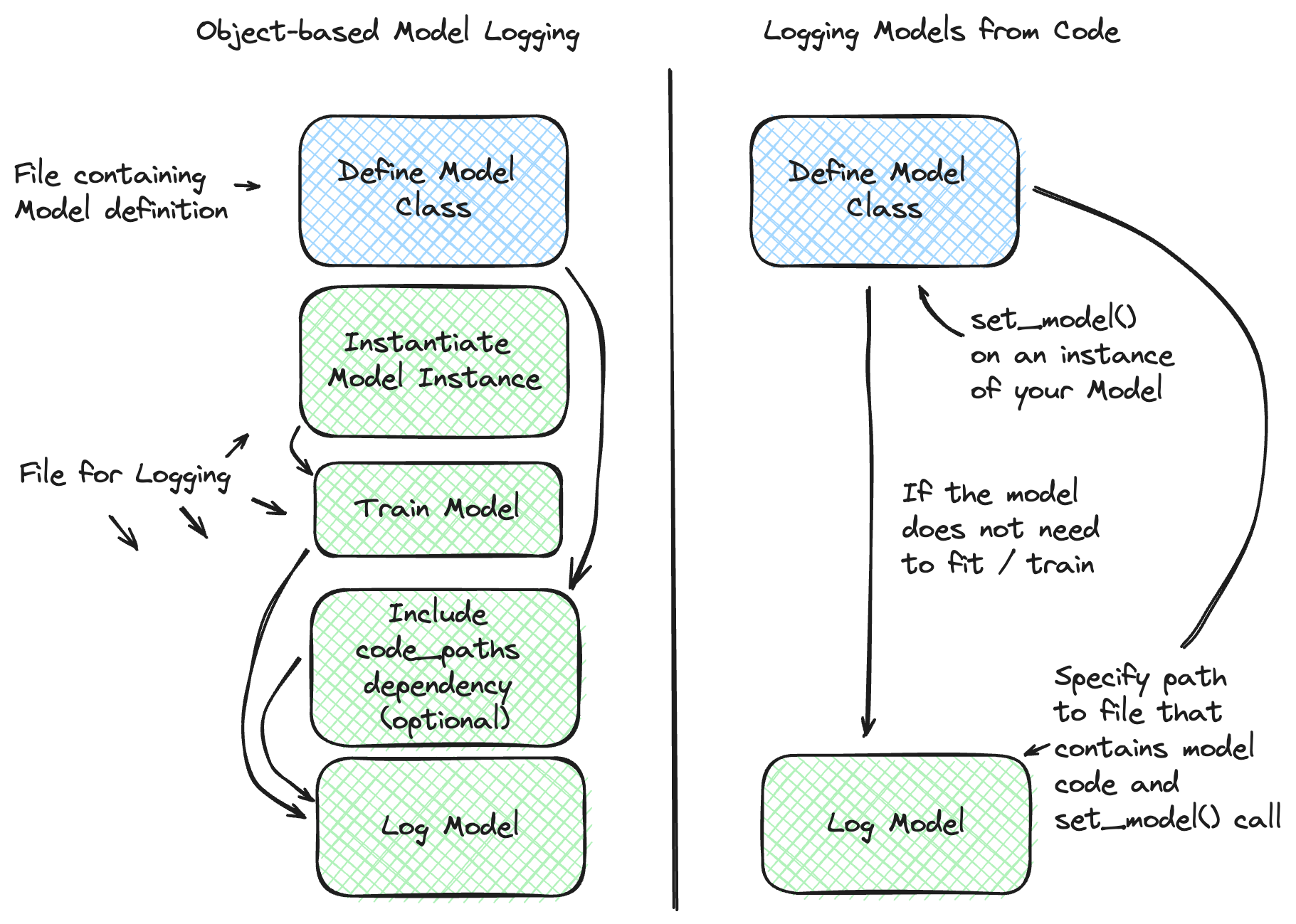
Models From Code
To learn more about the Models From Code feature, please visit the deep dive guide for more in-depth explanation and to see additional examples.
Note
The Models from Code feature is available in MLflow versions 2.12.2 and later. This feature is experimental and may change in future releases.
The Models from Code feature allows you to define and log models directly from a stand-alone python script. This feature is particularly useful when you want to
log models that can be effectively stored as a code representation (models that do not need optimized weights through training) or applications
that rely on external services (e.g., LangChain chains). Another benefit is that this approach entirely bypasses the use of the pickle or
cloudpickle modules within Python, which can carry security risks when loading untrusted models.
Note
This feature is only supported for LangChain, LlamaIndex, and PythonModel models.
In order to log a model from code, you can leverage the mlflow.models.set_model() API. This API allows you to define a model by specifying
an instance of the model class directly within the file where the model is defined. When logging such a model, a
file path is specified (instead of an object) that points to the Python file containing both the model class definition and the usage of the
set_model API applied on an instance of your custom model.
The figure below provides a comparison of the standard model logging process and the Models from Code feature for models that are eligible to be saved using the Models from Code feature:
For example, defining a model in a separate file named my_model.py:
import mlflow
from mlflow.models import set_model
class MyModel(mlflow.pyfunc.PythonModel):
def predict(self, context, model_input):
return model_input
# Define the custom PythonModel instance that will be used for inference
set_model(MyModel())
Note
The Models from code feature does not support capturing import statements that are from external file references. If you have dependencies that
are not captured via a pip install, dependencies will need to be included and resolved via appropriate absolute path import references from
using the code_paths feature.
For simplicity’s sake, it is recommended to encapsulate all of your required local dependencies for a model defined from code within the same
python script file due to limitations around code_paths dependency pathing resolution.
Tip
When defining a model from code and using the mlflow.models.set_model() API, the code that is defined in the script that is being logged
will be executed internally to ensure that it is valid code. If you have connections to external services within your script (e.g. you are connecting
to a GenAI service within LangChain), be aware that you will incur a connection request to that service when the model is being logged.
Then, logging the model from the file path in a different python script:
import mlflow
model_path = "my_model.py"
with mlflow.start_run():
model_info = mlflow.pyfunc.log_model(
python_model=model_path, # Define the model as the path to the Python file
artifact_path="my_model",
)
# Loading the model behaves exactly as if an instance of MyModel had been logged
my_model = mlflow.pyfunc.load_model(model_info.model_uri)
Warning
The mlflow.models.set_model() API is not threadsafe. Do not attempt to use this feature if you are logging models concurrently
from multiple threads. This fluent API utilizes a global active model state that has no consistency guarantees. If you are interested in threadsafe
logging APIs, please use the mlflow.client.MlflowClient APIs for logging models.
Built-In Model Flavors
MLflow provides several standard flavors that might be useful in your applications. Specifically, many of its deployment tools support these flavors, so you can export your own model in one of these flavors to benefit from all these tools:
Python Function (python_function)
The python_function model flavor serves as a default model interface for MLflow Python models.
Any MLflow Python model is expected to be loadable as a python_function model. This enables
other MLflow tools to work with any python model regardless of which persistence module or
framework was used to produce the model. This interoperability is very powerful because it allows
any Python model to be productionized in a variety of environments.
In addition, the python_function model flavor defines a generic filesystem model format for Python models and provides utilities for saving and loading models
to and from this format. The format is self-contained in the sense that it includes all the
information necessary to load and use a model. Dependencies are stored either directly with the
model or referenced via conda environment. This model format allows other tools to integrate
their models with MLflow.
How To Save Model As Python Function
Most python_function models are saved as part of other model flavors - for example, all mlflow
built-in flavors include the python_function flavor in the exported models. In addition, the
mlflow.pyfunc module defines functions for creating python_function models explicitly.
This module also includes utilities for creating custom Python models, which is a convenient way of
adding custom python code to ML models. For more information, see the custom Python models
documentation.
For information on how to store a custom model from a python script (models from code functionality), see the guide to models from code for the recommended approaches.
How To Load And Score Python Function Models
Loading Models
You can load python_function models in Python by using the mlflow.pyfunc.load_model() function. It is important
to note that load_model assumes all dependencies are already available and will not perform any checks or installations
of dependencies. For deployment options that handle dependencies, refer to the model deployment section.
Scoring Models
Once a model is loaded, it can be scored in two primary ways:
Synchronous Scoring The standard method for scoring is using the
predictmethod, which supports various input types and returns a scalar or collection based on the input data. The method signature is:predict(data: Union[pandas.Series, pandas.DataFrame, numpy.ndarray, csc_matrix, csr_matrix, List[Any], Dict[str, Any], str], params: Optional[Dict[str, Any]] = None) → Union[pandas.Series, pandas.DataFrame, numpy.ndarray, list, str]Synchronous Streaming Scoring
Note
predict_streamis a new interface that was added to MLflow in the 2.12.2 release. Previous versions of MLflow will not support this interface. In order to utilizepredict_streamin a custom Python Function Model, you must implement thepredict_streammethod in your model class and return a generator type.For models that support streaming data processing,
predict_streammethod is available. This method returns agenerator, which yields a stream of responses, allowing for efficient processing of large datasets or continuous data streams. Note that thepredict_streammethod is not available for all model types. The usage involves iterating over the generator to consume the responses:predict_stream(data: Any, params: Optional[Dict[str, Any]] = None) → GeneratorType
Demonstrating predict_stream()
Below is an example demonstrating how to define, save, load, and use a streamable model with the predict_stream() method:
import mlflow
import os
# Define a custom model that supports streaming
class StreamableModel(mlflow.pyfunc.PythonModel):
def predict(self, context, model_input, params=None):
# Regular predict method implementation (optional for this demo)
return "regular-predict-output"
def predict_stream(self, context, model_input, params=None):
# Yielding elements one at a time
for element in ["a", "b", "c", "d", "e"]:
yield element
# Save the model to a directory
tmp_path = "/tmp/test_model"
pyfunc_model_path = os.path.join(tmp_path, "pyfunc_model")
python_model = StreamableModel()
mlflow.pyfunc.save_model(path=pyfunc_model_path, python_model=python_model)
# Load the model
loaded_pyfunc_model = mlflow.pyfunc.load_model(model_uri=pyfunc_model_path)
# Use predict_stream to get a generator
stream_output = loaded_pyfunc_model.predict_stream("single-input")
# Consuming the generator using next
print(next(stream_output)) # Output: 'a'
print(next(stream_output)) # Output: 'b'
# Alternatively, consuming the generator using a for-loop
for response in stream_output:
print(response) # This will print 'c', 'd', 'e'
Python Function Model Interfaces
All PyFunc models will support pandas.DataFrame as an input. In addition to pandas.DataFrame, DL PyFunc models will also support tensor inputs in the form of numpy.ndarrays. To verify whether a model flavor supports tensor inputs, please check the flavor’s documentation.
For models with a column-based schema, inputs are typically provided in the form of a pandas.DataFrame. If a dictionary mapping column name to values is provided as input for schemas with named columns or if a python List or a numpy.ndarray is provided as input for schemas with unnamed columns, MLflow will cast the input to a DataFrame. Schema enforcement and casting with respect to the expected data types is performed against the DataFrame.
For models with a tensor-based schema, inputs are typically provided in the form of a numpy.ndarray or a dictionary mapping the tensor name to its np.ndarray value. Schema enforcement will check the provided input’s shape and type against the shape and type specified in the model’s schema and throw an error if they do not match.
For models where no schema is defined, no changes to the model inputs and outputs are made. MLflow will propagate any errors raised by the model if the model does not accept the provided input type.
The python environment that a PyFunc model is loaded into for prediction or inference may differ from the environment
in which it was trained. In the case of an environment mismatch, a warning message will be printed when calling
mlflow.pyfunc.load_model(). This warning statement will identify the packages that have a version mismatch
between those used during training and the current environment. In order to get the full dependencies of the
environment in which the model was trained, you can call mlflow.pyfunc.get_model_dependencies().
Furthermore, if you want to run model inference in the same environment used in model training, you can call
mlflow.pyfunc.spark_udf() with the env_manager argument set as “conda”. This will generate the environment
from the conda.yaml file, ensuring that the python UDF will execute with the exact package versions that were used
during training.
Some PyFunc models may accept model load configuration, which controls how the model is loaded and predictions computed. You can learn which configuration the model supports by inspecting the model’s flavor metadata:
model_info = mlflow.models.get_model_info(model_uri)
model_info.flavors[mlflow.pyfunc.FLAVOR_NAME][mlflow.pyfunc.MODEL_CONFIG]
Alternatively, you can load the PyFunc model and inspect the model_config property:
pyfunc_model = mlflow.pyfunc.load_model(model_uri)
pyfunc_model.model_config
Model configuration can be changed at loading time by indicating model_config parameter in the
mlflow.pyfunc.load_model() method:
pyfunc_model = mlflow.pyfunc.load_model(model_uri, model_config=dict(temperature=0.93))
When a model configuration value is changed, those values the configuration the model was saved with. Indicating an invalid model configuration key for a model results in that configuration being ignored. A warning is displayed mentioning the ignored entries.
Note
Model configuration vs parameters with default values in signatures: Use model configuration when you need to provide model publishers for a way to change how the model is loaded into memory and how predictions are computed for all the samples. For instance, a key like user_gpu. Model consumers are not able to change those values at predict time. Use parameters with default values in the signature to provide a users the ability to change how predictions are computed on each data sample.
R Function (crate)
The crate model flavor defines a generic model format for representing an arbitrary R prediction
function as an MLflow model using the crate function from the
carrier package. The prediction function is expected to take a dataframe as input and
produce a dataframe, a vector or a list with the predictions as output.
This flavor requires R to be installed in order to be used.
crate usage
For a minimal crate model, an example configuration for the predict function is:
library(mlflow)
library(carrier)
# Load iris dataset
data("iris")
# Learn simple linear regression model
model <- lm(Sepal.Width~Sepal.Length, data = iris)
# Define a crate model
# call package functions with an explicit :: namespace.
crate_model <- crate(
function(new_obs) stats::predict(model, data.frame("Sepal.Length" = new_obs)),
model = model
)
# log the model
model_path <- mlflow_log_model(model = crate_model, artifact_path = "iris_prediction")
# load the logged model and make a prediction
model_uri <- paste0(mlflow_get_run()$artifact_uri, "/iris_prediction")
mlflow_model <- mlflow_load_model(model_uri = model_uri,
flavor = NULL,
client = mlflow_client())
prediction <- mlflow_predict(model = mlflow_model, data = 5)
print(prediction)
H2O (h2o)
The h2o model flavor enables logging and loading H2O models.
The mlflow.h2o module defines save_model() and
log_model() methods in python, and
mlflow_save_model and
mlflow_log_model in R for saving H2O models in MLflow Model
format.
These methods produce MLflow Models with the python_function flavor, allowing you to load them
as generic Python functions for inference via mlflow.pyfunc.load_model().
This loaded PyFunc model can be scored with only DataFrame input. When you load
MLflow Models with the h2o flavor using mlflow.pyfunc.load_model(),
the h2o.init() method is
called. Therefore, the correct version of h2o(-py) must be installed in the loader’s
environment. You can customize the arguments given to
h2o.init() by modifying the
init entry of the persisted H2O model’s YAML configuration file: model.h2o/h2o.yaml.
Finally, you can use the mlflow.h2o.load_model() method to load MLflow Models with the
h2o flavor as H2O model objects.
For more information, see mlflow.h2o.
h2o pyfunc usage
For a minimal h2o model, here is an example of the pyfunc predict() method in a classification scenario :
import mlflow
import h2o
h2o.init()
from h2o.estimators.glm import H2OGeneralizedLinearEstimator
# import the prostate data
df = h2o.import_file(
"http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip"
)
# convert the columns to factors
df["CAPSULE"] = df["CAPSULE"].asfactor()
df["RACE"] = df["RACE"].asfactor()
df["DCAPS"] = df["DCAPS"].asfactor()
df["DPROS"] = df["DPROS"].asfactor()
# split the data
train, test, valid = df.split_frame(ratios=[0.7, 0.15])
# generate a GLM model
glm_classifier = H2OGeneralizedLinearEstimator(
family="binomial", lambda_=0, alpha=0.5, nfolds=5, compute_p_values=True
)
with mlflow.start_run():
glm_classifier.train(
y="CAPSULE", x=["AGE", "RACE", "VOL", "GLEASON"], training_frame=train
)
metrics = glm_classifier.model_performance()
metrics_to_track = ["MSE", "RMSE", "r2", "logloss"]
metrics_to_log = {
key: value
for key, value in metrics._metric_json.items()
if key in metrics_to_track
}
params = glm_classifier.params
mlflow.log_params(params)
mlflow.log_metrics(metrics_to_log)
model_info = mlflow.h2o.log_model(glm_classifier, artifact_path="h2o_model_info")
# load h2o model and make a prediction
h2o_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
test_df = test.as_data_frame()
predictions = h2o_pyfunc.predict(test_df)
print(predictions)
# it is also possible to load the model and predict using h2o methods on the h2o frame
# h2o_model = mlflow.h2o.load_model(model_info.model_uri)
# predictions = h2o_model.predict(test)
Keras (keras)
The keras model flavor enables logging and loading Keras models. It is available in both Python
and R clients. In R, you can save or log the model using
mlflow_save_model and mlflow_log_model.
These functions serialize Keras models as HDF5 files using the Keras library’s built-in
model persistence functions. You can use
mlflow_load_model function in R to load MLflow Models
with the keras flavor as Keras Model objects.
Keras pyfunc usage
For a minimal Sequential model, an example configuration for the pyfunc predict() method is:
import mlflow
import numpy as np
import pathlib
import shutil
from tensorflow import keras
mlflow.tensorflow.autolog()
X = np.array([-2, -1, 0, 1, 2, 1]).reshape(-1, 1)
y = np.array([0, 0, 1, 1, 1, 0])
model = keras.Sequential(
[
keras.Input(shape=(1,)),
keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.fit(X, y, batch_size=3, epochs=5, validation_split=0.2)
local_artifact_dir = "/tmp/mlflow/keras_model"
pathlib.Path(local_artifact_dir).mkdir(parents=True, exist_ok=True)
model_uri = f"runs:/{mlflow.last_active_run().info.run_id}/model"
keras_pyfunc = mlflow.pyfunc.load_model(
model_uri=model_uri, dst_path=local_artifact_dir
)
data = np.array([-4, 1, 0, 10, -2, 1]).reshape(-1, 1)
predictions = keras_pyfunc.predict(data)
shutil.rmtree(local_artifact_dir)
MLeap (mleap)
Warning
The mleap model flavor is deprecated as of MLflow 2.6.0 and will be removed in a future release.
The mleap model flavor supports saving Spark models in MLflow format using the
MLeap persistence mechanism. MLeap is an inference-optimized
format and execution engine for Spark models that does not depend on
SparkContext
to evaluate inputs.
Note
You can save Spark models in MLflow format with the mleap flavor by specifying the
sample_input argument of the mlflow.spark.save_model() or
mlflow.spark.log_model() method (recommended). For more details see Spark MLlib.
The mlflow.mleap module also
defines save_model() and
log_model() methods for saving MLeap models in MLflow format,
but these methods do not include the python_function flavor in the models they produce.
Similarly, mleap models can be saved in R with mlflow_save_model and loaded with mlflow_load_model, with
mlflow_save_model requiring sample_input to be specified as a
sample Spark dataframe containing input data to the model is required by MLeap for data schema
inference.
A companion module for loading MLflow Models with the MLeap flavor is available in the
mlflow/java package.
For more information, see mlflow.spark, mlflow.mleap, and the
MLeap documentation.
PyTorch (pytorch)
The pytorch model flavor enables logging and loading PyTorch models.
The mlflow.pytorch module defines utilities for saving and loading MLflow Models with the
pytorch flavor. You can use the mlflow.pytorch.save_model() and
mlflow.pytorch.log_model() methods to save PyTorch models in MLflow format; both of these
functions use the torch.save() method to
serialize PyTorch models. Additionally, you can use the mlflow.pytorch.load_model()
method to load MLflow Models with the pytorch flavor as PyTorch model objects. This loaded
PyFunc model can be scored with both DataFrame input and numpy array input. Finally, models
produced by mlflow.pytorch.save_model() and mlflow.pytorch.log_model() contain
the python_function flavor, allowing you to load them as generic Python functions for inference
via mlflow.pyfunc.load_model().
Note
When using the PyTorch flavor, if a GPU is available at prediction time, the default GPU will be used to run inference. To disable this behavior, users can use the MLFLOW_DEFAULT_PREDICTION_DEVICE or pass in a device with the device parameter for the predict function.
Note
In case of multi gpu training, ensure to save the model only with global rank 0 gpu. This avoids logging multiple copies of the same model.
PyTorch pyfunc usage
For a minimal PyTorch model, an example configuration for the pyfunc predict() method is:
import numpy as np
import mlflow
from mlflow.models import infer_signature
import torch
from torch import nn
net = nn.Linear(6, 1)
loss_function = nn.L1Loss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-4)
X = torch.randn(6)
y = torch.randn(1)
epochs = 5
for epoch in range(epochs):
optimizer.zero_grad()
outputs = net(X)
loss = loss_function(outputs, y)
loss.backward()
optimizer.step()
with mlflow.start_run() as run:
signature = infer_signature(X.numpy(), net(X).detach().numpy())
model_info = mlflow.pytorch.log_model(net, "model", signature=signature)
pytorch_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
predictions = pytorch_pyfunc.predict(torch.randn(6).numpy())
print(predictions)
For more information, see mlflow.pytorch.
Scikit-learn (sklearn)
The sklearn model flavor provides an easy-to-use interface for saving and loading scikit-learn
models. The mlflow.sklearn module defines
save_model() and
log_model() functions that save scikit-learn models in
MLflow format, using either Python’s pickle module (Pickle) or CloudPickle for model serialization.
These functions produce MLflow Models with the python_function flavor, allowing them to
be loaded as generic Python functions for inference via mlflow.pyfunc.load_model().
This loaded PyFunc model can only be scored with DataFrame input. Finally, you can use the
mlflow.sklearn.load_model() method to load MLflow Models with the sklearn flavor as
scikit-learn model objects.
Scikit-learn pyfunc usage
For a Scikit-learn LogisticRegression model, an example configuration for the pyfunc predict() method is:
import mlflow
from mlflow.models import infer_signature
import numpy as np
from sklearn.linear_model import LogisticRegression
with mlflow.start_run():
X = np.array([-2, -1, 0, 1, 2, 1]).reshape(-1, 1)
y = np.array([0, 0, 1, 1, 1, 0])
lr = LogisticRegression()
lr.fit(X, y)
signature = infer_signature(X, lr.predict(X))
model_info = mlflow.sklearn.log_model(
sk_model=lr, artifact_path="model", signature=signature
)
sklearn_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
data = np.array([-4, 1, 0, 10, -2, 1]).reshape(-1, 1)
predictions = sklearn_pyfunc.predict(data)
For more information, see mlflow.sklearn.
Spark MLlib (spark)
The spark model flavor enables exporting Spark MLlib models as MLflow Models.
The mlflow.spark module defines
save_model()to save a Spark MLlib model to a DBFS path.log_model()to upload a Spark MLlib model to the tracking server.mlflow.spark.load_model()to load MLflow Models with thesparkflavor as Spark MLlib pipelines.
MLflow Models produced by these functions contain the python_function flavor,
allowing you to load them as generic Python functions via mlflow.pyfunc.load_model().
This loaded PyFunc model can only be scored with DataFrame input.
When a model with the spark flavor is loaded as a Python function via
mlflow.pyfunc.load_model(), a new
SparkContext
is created for model inference; additionally, the function converts all Pandas DataFrame inputs to
Spark DataFrames before scoring. While this initialization overhead and format translation latency
is not ideal for high-performance use cases, it enables you to easily deploy any
MLlib PipelineModel to any production environment supported by MLflow
(SageMaker, AzureML, etc).
Spark MLlib pyfunc usage
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.linalg import Vectors
from pyspark.sql import SparkSession
import mlflow
# Prepare training data from a list of (label, features) tuples.
spark = SparkSession.builder.appName("LogisticRegressionExample").getOrCreate()
training = spark.createDataFrame(
[
(1.0, Vectors.dense([0.0, 1.1, 0.1])),
(0.0, Vectors.dense([2.0, 1.0, -1.0])),
(0.0, Vectors.dense([2.0, 1.3, 1.0])),
(1.0, Vectors.dense([0.0, 1.2, -0.5])),
],
["label", "features"],
)
# Create and fit a LogisticRegression instance
lr = LogisticRegression(maxIter=10, regParam=0.01)
lr_model = lr.fit(training)
# Serialize the Model
with mlflow.start_run():
model_info = mlflow.spark.log_model(lr_model, "spark-model")
# Load saved model
lr_model_saved = mlflow.pyfunc.load_model(model_info.model_uri)
# Make predictions on test data.
# The DataFrame used in the predict method must be a Pandas DataFrame
test = spark.createDataFrame(
[
(1.0, Vectors.dense([-1.0, 1.5, 1.3])),
(0.0, Vectors.dense([3.0, 2.0, -0.1])),
(1.0, Vectors.dense([0.0, 2.2, -1.5])),
],
["label", "features"],
).toPandas()
prediction = lr_model_saved.predict(test)
Note
Note that when the sample_input parameter is provided to log_model() or
save_model(), the Spark model is automatically saved as an mleap flavor
by invoking mlflow.mleap.add_to_model().
For example, the follow code block:
training_df = spark.createDataFrame([
(0, "a b c d e spark", 1.0),
(1, "b d", 0.0),
(2, "spark f g h", 1.0),
(3, "hadoop mapreduce", 0.0) ], ["id", "text", "label"])
tokenizer = Tokenizer(inputCol="text", outputCol="words")
hashingTF = HashingTF(inputCol=tokenizer.getOutputCol(), outputCol="features")
lr = LogisticRegression(maxIter=10, regParam=0.001)
pipeline = Pipeline(stages=[tokenizer, hashingTF, lr])
model = pipeline.fit(training_df)
mlflow.spark.log_model(model, "spark-model", sample_input=training_df)
results in the following directory structure logged to the MLflow Experiment:
# Directory written by with the addition of mlflow.mleap.add_to_model(model, "spark-model", training_df)
# Note the addition of the mleap directory
spark-model/
├── mleap
├── sparkml
├── MLmodel
├── conda.yaml
├── python_env.yaml
└── requirements.txt
For more information, see mlflow.mleap.
For more information, see mlflow.spark.
TensorFlow (tensorflow)
The simple example below shows how to log params and metrics in mlflow for a custom training loop
using low-level TensorFlow API. See tf-keras-example. for an example of mlflow and tf.keras models.
import numpy as np
import tensorflow as tf
import mlflow
x = np.linspace(-4, 4, num=512)
y = 3 * x + 10
# estimate w and b where y = w * x + b
learning_rate = 0.1
x_train = tf.Variable(x, trainable=False, dtype=tf.float32)
y_train = tf.Variable(y, trainable=False, dtype=tf.float32)
# initial values
w = tf.Variable(1.0)
b = tf.Variable(1.0)
with mlflow.start_run():
mlflow.log_param("learning_rate", learning_rate)
for i in range(1000):
with tf.GradientTape(persistent=True) as tape:
# calculate MSE = 0.5 * (y_predict - y_train)^2
y_predict = w * x_train + b
loss = 0.5 * tf.reduce_mean(tf.square(y_predict - y_train))
mlflow.log_metric("loss", value=loss.numpy(), step=i)
# Update the trainable variables
# w = w - learning_rate * gradient of loss function w.r.t. w
# b = b - learning_rate * gradient of loss function w.r.t. b
w.assign_sub(learning_rate * tape.gradient(loss, w))
b.assign_sub(learning_rate * tape.gradient(loss, b))
print(f"W = {w.numpy():.2f}, b = {b.numpy():.2f}")
ONNX (onnx)
The onnx model flavor enables logging of ONNX models in MLflow format via
the mlflow.onnx.save_model() and mlflow.onnx.log_model() methods. These
methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). This loaded PyFunc model can be scored with
both DataFrame input and numpy array input. The python_function representation of an MLflow
ONNX model uses the ONNX Runtime execution engine for
evaluation. Finally, you can use the mlflow.onnx.load_model() method to load MLflow
Models with the onnx flavor in native ONNX format.
For more information, see mlflow.onnx and http://onnx.ai/.
Warning
The default behavior for saving ONNX files is to use the ONNX save option save_as_external_data=True
in order to support model files that are in excess of 2GB. For edge deployments of small model files, this
may create issues. If you need to save a small model as a single file for such deployment considerations,
you can set the parameter save_as_external_data=False in either mlflow.onnx.save_model() or
mlflow.onnx.log_model() to force the serialization of the model as a small file. Note that if the
model is in excess of 2GB, saving as a single file will not work.
ONNX pyfunc usage example
For an ONNX model, an example configuration that uses pytorch to train a dummy model, converts it to ONNX, logs to mlflow and makes a prediction using pyfunc predict() method is:
import numpy as np
import mlflow
from mlflow.models import infer_signature
import onnx
import torch
from torch import nn
# define a torch model
net = nn.Linear(6, 1)
loss_function = nn.L1Loss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-4)
X = torch.randn(6)
y = torch.randn(1)
# run model training
epochs = 5
for epoch in range(epochs):
optimizer.zero_grad()
outputs = net(X)
loss = loss_function(outputs, y)
loss.backward()
optimizer.step()
# convert model to ONNX and load it
torch.onnx.export(net, X, "model.onnx")
onnx_model = onnx.load_model("model.onnx")
# log the model into a mlflow run
with mlflow.start_run():
signature = infer_signature(X.numpy(), net(X).detach().numpy())
model_info = mlflow.onnx.log_model(onnx_model, "model", signature=signature)
# load the logged model and make a prediction
onnx_pyfunc = mlflow.pyfunc.load_model(model_info.model_uri)
predictions = onnx_pyfunc.predict(X.numpy())
print(predictions)
XGBoost (xgboost)
The xgboost model flavor enables logging of XGBoost models
in MLflow format via the mlflow.xgboost.save_model() and mlflow.xgboost.log_model() methods in python and mlflow_save_model and mlflow_log_model in R respectively.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). This loaded PyFunc model can only be scored with DataFrame input.
You can also use the mlflow.xgboost.load_model()
method to load MLflow Models with the xgboost model flavor in native XGBoost format.
Note that the xgboost model flavor only supports an instance of xgboost.Booster,
not models that implement the scikit-learn API.
XGBoost pyfunc usage
The example below
Loads the IRIS dataset from
scikit-learnTrains an XGBoost Classifier
Logs the model and params using
mlflowLoads the logged model and makes predictions
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from xgboost import XGBClassifier
import mlflow
from mlflow.models import infer_signature
data = load_iris()
X_train, X_test, y_train, y_test = train_test_split(
data["data"], data["target"], test_size=0.2
)
xgb_classifier = XGBClassifier(
n_estimators=10,
max_depth=3,
learning_rate=1,
objective="binary:logistic",
random_state=123,
)
# log fitted model and XGBClassifier parameters
with mlflow.start_run():
xgb_classifier.fit(X_train, y_train)
clf_params = xgb_classifier.get_xgb_params()
mlflow.log_params(clf_params)
signature = infer_signature(X_train, xgb_classifier.predict(X_train))
model_info = mlflow.xgboost.log_model(
xgb_classifier, "iris-classifier", signature=signature
)
# Load saved model and make predictions
xgb_classifier_saved = mlflow.pyfunc.load_model(model_info.model_uri)
y_pred = xgb_classifier_saved.predict(X_test)
For more information, see mlflow.xgboost.
LightGBM (lightgbm)
The lightgbm model flavor enables logging of LightGBM models
in MLflow format via the mlflow.lightgbm.save_model() and mlflow.lightgbm.log_model() methods.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). You can also use the mlflow.lightgbm.load_model()
method to load MLflow Models with the lightgbm model flavor in native LightGBM format.
Note that the scikit-learn API for LightGBM is now supported. For more information, see mlflow.lightgbm.
LightGBM pyfunc usage
The example below
Loads the IRIS dataset from
scikit-learnTrains a LightGBM
LGBMClassifierLogs the model and feature importance’s using
mlflowLoads the logged model and makes predictions
from lightgbm import LGBMClassifier
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
import mlflow
from mlflow.models import infer_signature
data = load_iris()
# Remove special characters from feature names to be able to use them as keys for mlflow metrics
feature_names = [
name.replace(" ", "_").replace("(", "").replace(")", "")
for name in data["feature_names"]
]
X_train, X_test, y_train, y_test = train_test_split(
data["data"], data["target"], test_size=0.2
)
# create model instance
lgb_classifier = LGBMClassifier(
n_estimators=10,
max_depth=3,
learning_rate=1,
objective="binary:logistic",
random_state=123,
)
# Fit and save model and LGBMClassifier feature importances as mlflow metrics
with mlflow.start_run():
lgb_classifier.fit(X_train, y_train)
feature_importances = dict(zip(feature_names, lgb_classifier.feature_importances_))
feature_importance_metrics = {
f"feature_importance_{feature_name}": imp_value
for feature_name, imp_value in feature_importances.items()
}
mlflow.log_metrics(feature_importance_metrics)
signature = infer_signature(X_train, lgb_classifier.predict(X_train))
model_info = mlflow.lightgbm.log_model(
lgb_classifier, "iris-classifier", signature=signature
)
# Load saved model and make predictions
lgb_classifier_saved = mlflow.pyfunc.load_model(model_info.model_uri)
y_pred = lgb_classifier_saved.predict(X_test)
print(y_pred)
CatBoost (catboost)
The catboost model flavor enables logging of CatBoost models
in MLflow format via the mlflow.catboost.save_model() and mlflow.catboost.log_model() methods.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). You can also use the mlflow.catboost.load_model()
method to load MLflow Models with the catboost model flavor in native CatBoost format.
For more information, see mlflow.catboost.
CatBoost pyfunc usage
For a CatBoost Classifier model, an example configuration for the pyfunc predict() method is:
import mlflow
from mlflow.models import infer_signature
from catboost import CatBoostClassifier
from sklearn import datasets
# prepare data
X, y = datasets.load_wine(as_frame=False, return_X_y=True)
# train the model
model = CatBoostClassifier(
iterations=5,
loss_function="MultiClass",
allow_writing_files=False,
)
model.fit(X, y)
# create model signature
predictions = model.predict(X)
signature = infer_signature(X, predictions)
# log the model into a mlflow run
with mlflow.start_run():
model_info = mlflow.catboost.log_model(model, "model", signature=signature)
# load the logged model and make a prediction
catboost_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
print(catboost_pyfunc.predict(X[:5]))
Spacy(spaCy)
The spaCy model flavor enables logging of spaCy models in MLflow format via
the mlflow.spacy.save_model() and mlflow.spacy.log_model() methods. Additionally, these
methods add the python_function flavor to the MLflow Models that they produce, allowing the models to be
interpreted as generic Python functions for inference via mlflow.pyfunc.load_model().
This loaded PyFunc model can only be scored with DataFrame input. You can
also use the mlflow.spacy.load_model() method to load MLflow Models with the spacy model flavor
in native spaCy format.
For more information, see mlflow.spacy.
Spacy pyfunc usage
The example below shows how to train a Spacy TextCategorizer model, log the model artifact and metrics to the
mlflow tracking server and then load the saved model to make predictions. For this example, we will be using the
Polarity 2.0 dataset available in the nltk package. This dataset consists of 10000 positive and 10000 negative
short movie reviews.
First we convert the texts and sentiment labels (“pos” or “neg”) from NLTK native format to Spacy’s DocBin format:
import pandas as pd
import spacy
from nltk.corpus import movie_reviews
from spacy import Language
from spacy.tokens import DocBin
nltk.download("movie_reviews")
def get_sentences(sentiment_type: str) -> pd.DataFrame:
"""Reconstruct the sentences from the word lists for each review record for a specific ``sentiment_type``
as a pandas DataFrame with two columns: 'sentence' and 'sentiment'.
"""
file_ids = movie_reviews.fileids(sentiment_type)
sent_df = []
for file_id in file_ids:
sentence = " ".join(movie_reviews.words(file_id))
sent_df.append({"sentence": sentence, "sentiment": sentiment_type})
return pd.DataFrame(sent_df)
def convert(data_df: pd.DataFrame, target_file: str):
"""Convert a DataFrame with 'sentence' and 'sentiment' columns to a
spacy DocBin object and save it to 'target_file'.
"""
nlp = spacy.blank("en")
sentiment_labels = data_df.sentiment.unique()
spacy_doc = DocBin()
for _, row in data_df.iterrows():
sent_tokens = nlp.make_doc(row["sentence"])
# To train a Spacy TextCategorizer model, the label must be attached to the "cats" dictionary of the "Doc"
# object, e.g. {"pos": 1.0, "neg": 0.0} for a "pos" label.
for label in sentiment_labels:
sent_tokens.cats[label] = 1.0 if label == row["sentiment"] else 0.0
spacy_doc.add(sent_tokens)
spacy_doc.to_disk(target_file)
# Build a single DataFrame with both positive and negative reviews, one row per review
review_data = [get_sentences(sentiment_type) for sentiment_type in ("pos", "neg")]
review_data = pd.concat(review_data, axis=0)
# Split the DataFrame into a train and a dev set
train_df = review_data.groupby("sentiment", group_keys=False).apply(
lambda x: x.sample(frac=0.7, random_state=100)
)
dev_df = review_data.loc[review_data.index.difference(train_df.index), :]
# Save the train and dev data files to the current directory as "corpora.train" and "corpora.dev", respectively
convert(train_df, "corpora.train")
convert(dev_df, "corpora.dev")
To set up the training job, we first need to generate a configuration file as described in the Spacy Documentation
For simplicity, we will only use a TextCategorizer in the pipeline.
python -m spacy init config --pipeline textcat --lang en mlflow-textcat.cfg
Change the default train and dev paths in the config file to the current directory:
[paths]
- train = null
- dev = null
+ train = "."
+ dev = "."
In Spacy, the training loop is defined internally in Spacy’s code. Spacy provides a “logging” extension point where
we can use mlflow. To do this,
We have to define a function to write metrics / model input to
mlfowRegister it as a logger in
Spacy’s component registryChange the default console logger in the
Spacy’s configuration file (mlflow-textcat.cfg)
from typing import IO, Callable, Tuple, Dict, Any, Optional
import spacy
from spacy import Language
import mlflow
@spacy.registry.loggers("mlflow_logger.v1")
def mlflow_logger():
"""Returns a function, ``setup_logger`` that returns two functions:
* ``log_step`` is called internally by Spacy for every evaluation step. We can log the intermediate train and
validation scores to the mlflow tracking server here.
* ``finalize``: is called internally by Spacy after training is complete. We can log the model artifact to the
mlflow tracking server here.
"""
def setup_logger(
nlp: Language,
stdout: IO = sys.stdout,
stderr: IO = sys.stderr,
) -> Tuple[Callable, Callable]:
def log_step(info: Optional[Dict[str, Any]]):
if info:
step = info["step"]
score = info["score"]
metrics = {}
for pipe_name in nlp.pipe_names:
loss = info["losses"][pipe_name]
metrics[f"{pipe_name}_loss"] = loss
metrics[f"{pipe_name}_score"] = score
mlflow.log_metrics(metrics, step=step)
def finalize():
uri = mlflow.spacy.log_model(nlp, "mlflow_textcat_example")
mlflow.end_run()
return log_step, finalize
return setup_logger
Check the spacy-loggers library <https://pypi.org/project/spacy-loggers/> _ for a more complete implementation.
Point to our mlflow logger in Spacy configuration file. For this example, we will lower the number of training steps
and eval frequency:
[training.logger]
- @loggers = "spacy.ConsoleLogger.v1"
- dev = null
+ @loggers = "mlflow_logger.v1"
[training]
- max_steps = 20000
- eval_frequency = 100
+ max_steps = 100
+ eval_frequency = 10
Train our model:
from spacy.cli.train import train as spacy_train
spacy_train("mlflow-textcat.cfg")
To make predictions, we load the saved model from the last run:
from mlflow import MlflowClient
# look up the last run info from mlflow
client = MlflowClient()
last_run = client.search_runs(experiment_ids=["0"], max_results=1)[0]
# We need to append the spacy model directory name to the artifact uri
spacy_model = mlflow.pyfunc.load_model(
f"{last_run.info.artifact_uri}/mlflow_textcat_example"
)
predictions_in = dev_df.loc[:, ["sentence"]]
predictions_out = spacy_model.predict(predictions_in).squeeze().tolist()
predicted_labels = [
"pos" if row["pos"] > row["neg"] else "neg" for row in predictions_out
]
print(dev_df.assign(predicted_sentiment=predicted_labels))
Fastai(fastai)
The fastai model flavor enables logging of fastai Learner models in MLflow format via
the mlflow.fastai.save_model() and mlflow.fastai.log_model() methods. Additionally, these
methods add the python_function flavor to the MLflow Models that they produce, allowing the models to be
interpreted as generic Python functions for inference via mlflow.pyfunc.load_model(). This loaded PyFunc model can
only be scored with DataFrame input. You can also use the mlflow.fastai.load_model() method to
load MLflow Models with the fastai model flavor in native fastai format.
The interface for utilizing a fastai model loaded as a pyfunc type for generating predictions uses a
Pandas DataFrame argument.
This example runs the fastai tabular tutorial,
logs the experiments, saves the model in fastai format and loads the model to get predictions
using a fastai data loader:
from fastai.data.external import URLs, untar_data
from fastai.tabular.core import Categorify, FillMissing, Normalize, TabularPandas
from fastai.tabular.data import TabularDataLoaders
from fastai.tabular.learner import tabular_learner
from fastai.data.transforms import RandomSplitter
from fastai.metrics import accuracy
from fastcore.basics import range_of
import pandas as pd
import mlflow
import mlflow.fastai
def print_auto_logged_info(r):
tags = {k: v for k, v in r.data.tags.items() if not k.startswith("mlflow.")}
artifacts = [
f.path for f in mlflow.MlflowClient().list_artifacts(r.info.run_id, "model")
]
print(f"run_id: {r.info.run_id}")
print(f"artifacts: {artifacts}")
print(f"params: {r.data.params}")
print(f"metrics: {r.data.metrics}")
print(f"tags: {tags}")
def main(epochs=5, learning_rate=0.01):
path = untar_data(URLs.ADULT_SAMPLE)
path.ls()
df = pd.read_csv(path / "adult.csv")
dls = TabularDataLoaders.from_csv(
path / "adult.csv",
path=path,
y_names="salary",
cat_names=[
"workclass",
"education",
"marital-status",
"occupation",
"relationship",
"race",
],
cont_names=["age", "fnlwgt", "education-num"],
procs=[Categorify, FillMissing, Normalize],
)
splits = RandomSplitter(valid_pct=0.2)(range_of(df))
to = TabularPandas(
df,
procs=[Categorify, FillMissing, Normalize],
cat_names=[
"workclass",
"education",
"marital-status",
"occupation",
"relationship",
"race",
],
cont_names=["age", "fnlwgt", "education-num"],
y_names="salary",
splits=splits,
)
dls = to.dataloaders(bs=64)
model = tabular_learner(dls, metrics=accuracy)
mlflow.fastai.autolog()
with mlflow.start_run() as run:
model.fit(5, 0.01)
mlflow.fastai.log_model(model, "model")
print_auto_logged_info(mlflow.get_run(run_id=run.info.run_id))
model_uri = f"runs:/{run.info.run_id}/model"
loaded_model = mlflow.fastai.load_model(model_uri)
test_df = df.copy()
test_df.drop(["salary"], axis=1, inplace=True)
dl = learn.dls.test_dl(test_df)
predictions, _ = loaded_model.get_preds(dl=dl)
px = pd.DataFrame(predictions).astype("float")
px.head(5)
main()
Output (Pandas DataFrame):
Index |
Probability of first class |
Probability of second class |
|---|---|---|
0 |
0.545088 |
0.454912 |
1 |
0.503172 |
0.496828 |
2 |
0.962663 |
0.037337 |
3 |
0.206107 |
0.793893 |
4 |
0.807599 |
0.192401 |
Alternatively, when using the python_function flavor, get predictions from a DataFrame.
from fastai.data.external import URLs, untar_data
from fastai.tabular.core import Categorify, FillMissing, Normalize, TabularPandas
from fastai.tabular.data import TabularDataLoaders
from fastai.tabular.learner import tabular_learner
from fastai.data.transforms import RandomSplitter
from fastai.metrics import accuracy
from fastcore.basics import range_of
import pandas as pd
import mlflow
import mlflow.fastai
model_uri = ...
path = untar_data(URLs.ADULT_SAMPLE)
df = pd.read_csv(path / "adult.csv")
test_df = df.copy()
test_df.drop(["salary"], axis=1, inplace=True)
loaded_model = mlflow.pyfunc.load_model(model_uri)
loaded_model.predict(test_df)
Output (Pandas DataFrame):
Index |
Probability of first class, Probability of second class |
|---|---|
0 |
[0.5450878, 0.45491222] |
1 |
[0.50317234, 0.49682766] |
2 |
[0.9626626, 0.037337445] |
3 |
[0.20610662, 0.7938934] |
4 |
[0.8075987, 0.19240129] |
For more information, see mlflow.fastai.
Statsmodels (statsmodels)
The statsmodels model flavor enables logging of Statsmodels models in MLflow format via the mlflow.statsmodels.save_model()
and mlflow.statsmodels.log_model() methods.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). This loaded PyFunc model can only be scored with DataFrame input.
You can also use the mlflow.statsmodels.load_model()
method to load MLflow Models with the statsmodels model flavor in native statsmodels format.
As for now, automatic logging is restricted to parameters, metrics and models generated by a call to fit
on a statsmodels model.
Statsmodels pyfunc usage
The following 2 examples illustrate usage of a basic regression model (OLS) and an ARIMA time series model from the following statsmodels apis : statsmodels.formula.api and statsmodels.tsa.api
For a minimal statsmodels regression model, here is an example of the pyfunc predict() method :
import mlflow
import pandas as pd
from sklearn.datasets import load_diabetes
import statsmodels.formula.api as smf
# load the diabetes dataset from sklearn
diabetes = load_diabetes()
# create X and y dataframes for the features and target
X = pd.DataFrame(data=diabetes.data, columns=diabetes.feature_names)
y = pd.DataFrame(data=diabetes.target, columns=["target"])
# concatenate X and y dataframes
df = pd.concat([X, y], axis=1)
# create the linear regression model (ordinary least squares)
model = smf.ols(
formula="target ~ age + sex + bmi + bp + s1 + s2 + s3 + s4 + s5 + s6", data=df
)
mlflow.statsmodels.autolog(
log_models=True,
disable=False,
exclusive=False,
disable_for_unsupported_versions=False,
silent=False,
registered_model_name=None,
)
with mlflow.start_run():
res = model.fit(method="pinv", use_t=True)
model_info = mlflow.statsmodels.log_model(res, artifact_path="OLS_model")
# load the pyfunc model
statsmodels_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
# generate predictions
predictions = statsmodels_pyfunc.predict(X)
print(predictions)
For a minimal time series ARIMA model, here is an example of the pyfunc predict() method :
import mlflow
import numpy as np
import pandas as pd
from statsmodels.tsa.arima.model import ARIMA
# create a time series dataset with seasonality
np.random.seed(0)
# generate a time index with a daily frequency
dates = pd.date_range(start="2022-12-01", end="2023-12-01", freq="D")
# generate the seasonal component (weekly)
seasonality = np.sin(np.arange(len(dates)) * (2 * np.pi / 365.25) * 7)
# generate the trend component
trend = np.linspace(-5, 5, len(dates)) + 2 * np.sin(
np.arange(len(dates)) * (2 * np.pi / 365.25) * 0.1
)
# generate the residual component
residuals = np.random.normal(0, 1, len(dates))
# generate the final time series by adding the components
time_series = seasonality + trend + residuals
# create a dataframe from the time series
data = pd.DataFrame({"date": dates, "value": time_series})
data.set_index("date", inplace=True)
order = (1, 0, 0)
# create the ARIMA model
model = ARIMA(data, order=order)
mlflow.statsmodels.autolog(
log_models=True,
disable=False,
exclusive=False,
disable_for_unsupported_versions=False,
silent=False,
registered_model_name=None,
)
with mlflow.start_run():
res = model.fit()
mlflow.log_params(
{
"order": order,
"trend": model.trend,
"seasonal_order": model.seasonal_order,
}
)
mlflow.log_params(res.params)
mlflow.log_metric("aic", res.aic)
mlflow.log_metric("bic", res.bic)
model_info = mlflow.statsmodels.log_model(res, artifact_path="ARIMA_model")
# load the pyfunc model
statsmodels_pyfunc = mlflow.pyfunc.load_model(model_uri=model_info.model_uri)
# prediction dataframes for a TimeSeriesModel must have exactly one row and include columns called start and end
start = pd.to_datetime("2024-01-01")
end = pd.to_datetime("2024-01-07")
# generate predictions
prediction_data = pd.DataFrame({"start": start, "end": end}, index=[0])
predictions = statsmodels_pyfunc.predict(prediction_data)
print(predictions)
For more information, see mlflow.statsmodels.
Prophet (prophet)
The prophet model flavor enables logging of Prophet models in MLflow format via the mlflow.prophet.save_model()
and mlflow.prophet.log_model() methods.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). This loaded PyFunc model can only be scored with DataFrame input.
You can also use the mlflow.prophet.load_model()
method to load MLflow Models with the prophet model flavor in native prophet format.
Prophet pyfunc usage
This example uses a time series dataset from Prophet’s GitHub repository, containing log number of daily views to Peyton Manning’s Wikipedia page for several years. A sample of the dataset is as follows:
ds |
y |
|---|---|
2007-12-10 |
9.59076113897809 |
2007-12-11 |
8.51959031601596 |
2007-12-12 |
8.18367658262066 |
2007-12-13 |
8.07246736935477 |
import numpy as np
import pandas as pd
from prophet import Prophet, serialize
from prophet.diagnostics import cross_validation, performance_metrics
import mlflow
from mlflow.models import infer_signature
# URL to the dataset
SOURCE_DATA = "https://raw.githubusercontent.com/facebook/prophet/main/examples/example_wp_log_peyton_manning.csv"
np.random.seed(12345)
def extract_params(pr_model):
params = {attr: getattr(pr_model, attr) for attr in serialize.SIMPLE_ATTRIBUTES}
return {k: v for k, v in params.items() if isinstance(v, (int, float, str, bool))}
# Load the training data
train_df = pd.read_csv(SOURCE_DATA)
# Create a "test" DataFrame with the "ds" column containing 10 days after the end date in train_df
test_dates = pd.date_range(start="2016-01-21", end="2016-01-31", freq="D")
test_df = pd.DataFrame({"ds": test_dates})
# Initialize Prophet model with specific parameters
prophet_model = Prophet(changepoint_prior_scale=0.5, uncertainty_samples=7)
with mlflow.start_run():
# Fit the model on the training data
prophet_model.fit(train_df)
# Extract and log model parameters
params = extract_params(prophet_model)
mlflow.log_params(params)
# Perform cross-validation
cv_results = cross_validation(
prophet_model,
initial="900 days",
period="30 days",
horizon="30 days",
parallel="threads",
disable_tqdm=True,
)
# Calculate and log performance metrics
cv_metrics = performance_metrics(cv_results, metrics=["mse", "rmse", "mape"])
average_metrics = cv_metrics.drop(columns=["horizon"]).mean(axis=0).to_dict()
mlflow.log_metrics(average_metrics)
# Generate predictions and infer model signature
train = prophet_model.history
# Log the Prophet model with MLflow
model_info = mlflow.prophet.log_model(
prophet_model,
artifact_path="prophet_model",
input_example=train[["ds"]].head(10),
)
# Load the saved model as a pyfunc
prophet_model_saved = mlflow.pyfunc.load_model(model_info.model_uri)
# Generate predictions for the test set
predictions = prophet_model_saved.predict(test_df)
# Truncate and display the forecast if needed
forecast = predictions[["ds", "yhat"]]
print(f"forecast:\n{forecast.head(5)}")
Output (Pandas DataFrame):
Index |
ds |
yhat |
yhat_upper |
yhat_lower |
|---|---|---|---|---|
0 |
2016-01-21 |
8.526513 |
8.827397 |
8.328563 |
1 |
2016-01-22 |
8.541355 |
9.434994 |
8.112758 |
2 |
2016-01-23 |
8.308332 |
8.633746 |
8.201323 |
3 |
2016-01-24 |
8.676326 |
9.534593 |
8.020874 |
4 |
2016-01-25 |
8.983457 |
9.430136 |
8.121798 |
For more information, see mlflow.prophet.
Pmdarima (pmdarima)
The pmdarima model flavor enables logging of pmdarima models in MLflow
format via the mlflow.pmdarima.save_model() and mlflow.pmdarima.log_model() methods.
These methods also add the python_function flavor to the MLflow Models that they produce, allowing the
model to be interpreted as generic Python functions for inference via mlflow.pyfunc.load_model().
This loaded PyFunc model can only be scored with a DataFrame input.
You can also use the mlflow.pmdarima.load_model() method to load MLflow Models with the pmdarima
model flavor in native pmdarima formats.
The interface for utilizing a pmdarima model loaded as a pyfunc type for generating forecast predictions uses
a single-row Pandas DataFrame configuration argument. The following columns in this configuration
Pandas DataFrame are supported:
n_periods(required) - specifies the number of future periods to generate starting from the last datetime valueof the training dataset, utilizing the frequency of the input training series when the model was trained. (for example, if the training data series elements represent one value per hour, in order to forecast 3 days of future data, set the column
n_periodsto72.
X(optional) - exogenous regressor values (only supported in pmdarima version >= 1.8.0) a 2D array of values forfuture time period events. For more information, read the underlying library explanation.
return_conf_int(optional) - a boolean (Default:False) for whether to return confidence interval values.See above note.
alpha(optional) - the significance value for calculating confidence intervals. (Default:0.05)
An example configuration for the pyfunc predict of a pmdarima model is shown below, with a future period
prediction count of 100, a confidence interval calculation generation, no exogenous regressor elements, and a default
alpha of 0.05:
Index |
n_periods |
return_conf_int |
|---|---|---|
0 |
100 |
True |
Warning
The Pandas DataFrame passed to a pmdarima pyfunc flavor must only contain 1 row.
Note
When predicting a pmdarima flavor, the predict method’s DataFrame configuration column
return_conf_int’s value controls the output format. When the column’s value is set to False or None
(which is the default if this column is not supplied in the configuration DataFrame), the schema of the
returned Pandas DataFrame is a single column: ["yhat"]. When set to True, the schema of the returned
DataFrame is: ["yhat", "yhat_lower", "yhat_upper"] with the respective lower (yhat_lower) and
upper (yhat_upper) confidence intervals added to the forecast predictions (yhat).
Example usage of pmdarima artifact loaded as a pyfunc with confidence intervals calculated:
import pmdarima
import mlflow
import pandas as pd
data = pmdarima.datasets.load_airpassengers()
with mlflow.start_run():
model = pmdarima.auto_arima(data, seasonal=True)
mlflow.pmdarima.save_model(model, "/tmp/model.pmd")
loaded_pyfunc = mlflow.pyfunc.load_model("/tmp/model.pmd")
prediction_conf = pd.DataFrame(
[{"n_periods": 4, "return_conf_int": True, "alpha": 0.1}]
)
predictions = loaded_pyfunc.predict(prediction_conf)
Output (Pandas DataFrame):
Index |
yhat |
yhat_lower |
yhat_upper |
|---|---|---|---|
0 |
467.573731 |
423.30995 |
511.83751 |
1 |
490.494467 |
416.17449 |
564.81444 |
2 |
509.138684 |
420.56255 |
597.71117 |
3 |
492.554714 |
397.30634 |
587.80309 |
Warning
Signature logging for pmdarima will not function correctly if return_conf_int is set to True from
a non-pyfunc artifact. The output of the native ARIMA.predict() when returning confidence intervals is not
a recognized signature type.
OpenAI (openai) (Experimental)
The full guide, including tutorials and detailed documentation for using the openai flavor can be viewed here.
LangChain (langchain) (Experimental)
The full guide, including tutorials and detailed documentation for using the langchain flavor can be viewed here.
John Snow Labs (johnsnowlabs) (Experimental)
Attention
The johnsnowlabs flavor is in active development and is marked as Experimental. Public APIs may change and new features are
subject to be added as additional functionality is brought to the flavor.
The johnsnowlabs model flavor gives you access to 20.000+ state-of-the-art enterprise NLP models in 200+ languages for medical, finance, legal and many more domains.
You can use mlflow.johnsnowlabs.log_model() to log and export your model as
mlflow.pyfunc.PyFuncModel.
This enables you to integrate any John Snow Labs model
into the MLflow framework. You can easily deploy your models for inference with MLflows serve functionalities.
Models are interpreted as a generic Python function for inference via mlflow.pyfunc.load_model().
You can also use the mlflow.johnsnowlabs.load_model() function to load a saved or logged MLflow
Model with the johnsnowlabs flavor from an stored artifact.
Features include: LLM’s, Text Summarization, Question Answering, Named Entity Recognition, Relation Extraction, Sentiment Analysis, Spell Checking, Image Classification, Automatic Speech Recognition and much more, powered by the latest Transformer Architectures. The models are provided by John Snow Labs and requires a John Snow Labs Enterprise NLP License. You can reach out to us for a research or industry license.
Example: Export a John Snow Labs to MLflow format
import json
import os
import pandas as pd
from johnsnowlabs import nlp
import mlflow
from mlflow.pyfunc import spark_udf
# 1) Write your raw license.json string into the 'JOHNSNOWLABS_LICENSE_JSON' env variable for MLflow
creds = {
"AWS_ACCESS_KEY_ID": "...",
"AWS_SECRET_ACCESS_KEY": "...",
"SPARK_NLP_LICENSE": "...",
"SECRET": "...",
}
os.environ["JOHNSNOWLABS_LICENSE_JSON"] = json.dumps(creds)
# 2) Install enterprise libraries
nlp.install()
# 3) Start a Spark session with enterprise libraries
spark = nlp.start()
# 4) Load a model and test it
nlu_model = "en.classify.bert_sequence.covid_sentiment"
model_save_path = "my_model"
johnsnowlabs_model = nlp.load(nlu_model)
johnsnowlabs_model.predict(["I hate COVID,", "I love COVID"])
# 5) Export model with pyfunc and johnsnowlabs flavors
with mlflow.start_run():
model_info = mlflow.johnsnowlabs.log_model(johnsnowlabs_model, model_save_path)
# 6) Load model with johnsnowlabs flavor
mlflow.johnsnowlabs.load_model(model_info.model_uri)
# 7) Load model with pyfunc flavor
mlflow.pyfunc.load_model(model_save_path)
pandas_df = pd.DataFrame({"text": ["Hello World"]})
spark_df = spark.createDataFrame(pandas_df).coalesce(1)
pyfunc_udf = spark_udf(
spark=spark,
model_uri=model_save_path,
env_manager="virtualenv",
result_type="string",
)
new_df = spark_df.withColumn("prediction", pyfunc_udf(*pandas_df.columns))
# 9) You can now use the mlflow models serve command to serve the model see next section
# 10) You can also use x command to deploy model inside of a container see next section
To deploy the John Snow Labs model as a container
Start the Docker Container
docker run -p 5001:8080 -e JOHNSNOWLABS_LICENSE_JSON=your_json_string "mlflow-pyfunc"
Query server
curl http://127.0.0.1:5001/invocations -H 'Content-Type: application/json' -d '{
"dataframe_split": {
"columns": ["text"],
"data": [["I hate covid"], ["I love covid"]]
}
}'
To deploy the John Snow Labs model without a container
Export env variable and start server
export JOHNSNOWLABS_LICENSE_JSON=your_json_string
mlflow models serve -m <model_uri>
Query server
curl http://127.0.0.1:5000/invocations -H 'Content-Type: application/json' -d '{
"dataframe_split": {
"columns": ["text"],
"data": [["I hate covid"], ["I love covid"]]
}
}'
Diviner (diviner)
The diviner model flavor enables logging of
diviner models in MLflow format via the
mlflow.diviner.save_model() and mlflow.diviner.log_model() methods. These methods also add the
python_function flavor to the MLflow Models that they produce, allowing the model to be interpreted as generic
Python functions for inference via mlflow.pyfunc.load_model().
This loaded PyFunc model can only be scored with a DataFrame input.
You can also use the mlflow.diviner.load_model() method to load MLflow Models with the diviner
model flavor in native diviner formats.
Diviner Types
Diviner is a library that provides an orchestration framework for performing time series forecasting on groups of
related series. Forecasting in diviner is accomplished through wrapping popular open source libraries such as
prophet and pmdarima. The diviner
library offers a simplified set of APIs to simultaneously generate distinct time series forecasts for multiple data
groupings using a single input DataFrame and a unified high-level API.
Metrics and Parameters logging for Diviner
Unlike other flavors that are supported in MLflow, Diviner has the concept of grouped models. As a collection of many
(perhaps thousands) of individual forecasting models, the burden to the tracking server to log individual metrics
and parameters for each of these models is significant. For this reason, metrics and parameters are exposed for
retrieval from Diviner’s APIs as Pandas DataFrames, rather than discrete primitive values.
To illustrate, let us assume we are forecasting hourly electricity consumption from major cities around the world. A sample of our input data looks like this:
country |
city |
datetime |
watts |
|---|---|---|---|
US |
NewYork |
2022-03-01 00:01:00 |
23568.9 |
US |
NewYork |
2022-03-01 00:02:00 |
22331.7 |
US |
Boston |
2022-03-01 00:01:00 |
14220.1 |
US |
Boston |
2022-03-01 00:02:00 |
14183.4 |
CA |
Toronto |
2022-03-01 00:01:00 |
18562.2 |
CA |
Toronto |
2022-03-01 00:02:00 |
17681.6 |
MX |
MexicoCity |
2022-03-01 00:01:00 |
19946.8 |
MX |
MexicoCity |
2022-03-01 00:02:00 |
19444.0 |
If we were to fit a model on this data, supplying the grouping keys as:
grouping_keys = ["country", "city"]
We will have a model generated for each of the grouping keys that have been supplied:
[("US", "NewYork"), ("US", "Boston"), ("CA", "Toronto"), ("MX", "MexicoCity")]
With a model constructed for each of these, entering each of their metrics and parameters wouldn’t be an issue for the MLflow tracking server. What would become a problem, however, is if we modeled each major city on the planet and ran this forecasting scenario every day. If we were to adhere to the conditions of the World Bank, that would mean just over 10,000 models as of 2022. After a mere few weeks of running this forecasting every day we would have a very large metrics table.
To eliminate this issue for large-scale forecasting, the metrics and parameters for diviner are extracted as a
grouping key indexed Pandas DataFrame, as shown below for example (float values truncated for visibility):
grouping_key_columns |
country |
city |
mse |
rmse |
mae |
mape |
mdape |
smape |
|---|---|---|---|---|---|---|---|---|
“(‘country’, ‘city’)” |
CA |
Toronto |
8276851.6 |
2801.7 |
2417.7 |
0.16 |
0.16 |
0.159 |
“(‘country’, ‘city’)” |
MX |
MexicoCity |
3548872.4 |
1833.8 |
1584.5 |
0.15 |
0.16 |
0.159 |
“(‘country’, ‘city’)” |
US |
NewYork |
3167846.4 |
1732.4 |
1498.2 |
0.15 |
0.16 |
0.158 |
“(‘country’, ‘city’)” |
US |
Boston |
14082666.4 |
3653.2 |
3156.2 |
0.15 |
0.16 |
0.159 |
There are two recommended means of logging the metrics and parameters from a diviner model :
Writing the DataFrames to local storage and using
mlflow.log_artifacts()
import os
import mlflow
import tempfile
with tempfile.TemporaryDirectory() as tmpdir:
params = model.extract_model_params()
metrics = model.cross_validate_and_score(
horizon="72 hours",
period="240 hours",
initial="480 hours",
parallel="threads",
rolling_window=0.1,
monthly=False,
)
params.to_csv(f"{tmpdir}/params.csv", index=False, header=True)
metrics.to_csv(f"{tmpdir}/metrics.csv", index=False, header=True)
mlflow.log_artifacts(tmpdir, artifact_path="data")
Writing directly as a JSON artifact using
mlflow.log_dict()
Note
The parameters extract from diviner models may require casting (or dropping of columns) if using the
pd.DataFrame.to_dict() approach due to the inability of this method to serialize objects.
import mlflow
params = model.extract_model_params()
metrics = model.cross_validate_and_score(
horizon="72 hours",
period="240 hours",
initial="480 hours",
parallel="threads",
rolling_window=0.1,
monthly=False,
)
params["t_scale"] = params["t_scale"].astype(str)
params["start"] = params["start"].astype(str)
params = params.drop("stan_backend", axis=1)
mlflow.log_dict(params.to_dict(), "params.json")
mlflow.log_dict(metrics.to_dict(), "metrics.json")
Logging of the model artifact is shown in the pyfunc example below.
Diviner pyfunc usage
The MLflow Diviner flavor includes an implementation of the pyfunc interface for Diviner models. To control
prediction behavior, you can specify configuration arguments in the first row of a Pandas DataFrame input.
As this configuration is dependent upon the underlying model type (i.e., the diviner.GroupedProphet.forecast()
method has a different signature than does diviner.GroupedPmdarima.predict()), the Diviner pyfunc implementation
attempts to coerce arguments to the types expected by the underlying model.
Note
Diviner models support both “full group” and “partial group” forecasting. If a column named "groups" is present
in the configuration DataFrame submitted to the pyfunc flavor, the grouping key values in the first row
will be used to generate a subset of forecast predictions. This functionality removes the need to filter a subset
from the full output of all groups forecasts if the results of only a few (or one) groups are needed.
For a GroupedPmdarima model, an example configuration for the pyfunc predict() method is:
import mlflow
import pandas as pd
from pmdarima.arima.auto import AutoARIMA
from diviner import GroupedPmdarima
with mlflow.start_run():
base_model = AutoARIMA(out_of_sample_size=96, maxiter=200)
model = GroupedPmdarima(model_template=base_model).fit(
df=df,
group_key_columns=["country", "city"],
y_col="watts",
datetime_col="datetime",
silence_warnings=True,
)
mlflow.diviner.save_model(diviner_model=model, path="/tmp/diviner_model")
diviner_pyfunc = mlflow.pyfunc.load_model(model_uri="/tmp/diviner_model")
predict_conf = pd.DataFrame(
{
"n_periods": 120,
"groups": [
("US", "NewYork"),
("CA", "Toronto"),
("MX", "MexicoCity"),
], # NB: List of tuples required.
"predict_col": "wattage_forecast",
"alpha": 0.1,
"return_conf_int": True,
"on_error": "warn",
},
index=[0],
)
subset_forecasts = diviner_pyfunc.predict(predict_conf)
Note
There are several instances in which a configuration DataFrame submitted to the pyfunc predict() method
will cause an MlflowException to be raised:
If neither
horizonorn_periodsare provided.The value of
n_periodsorhorizonis not an integer.If the model is of type
GroupedProphet,frequencyas a string type must be provided.If both
horizonandn_periodsare provided with different values.
Transformers (transformers) (Experimental)
The full guide, including tutorials and detailed documentation for using the transformers flavor is available at this location.
SentenceTransformers (sentence_transformers) (Experimental)
Attention
The sentence_transformers flavor is in active development and is marked as Experimental. Public APIs may change and new
features are subject to be added as additional functionality is brought to the flavor.
The sentence_transformers model flavor enables logging of
sentence-transformers models in MLflow format via
the mlflow.sentence_transformers.save_model() and mlflow.sentence_transformers.log_model() functions.
Use of these functions also adds the python_function flavor to the MLflow Models that they produce, allowing the model to be
interpreted as a generic Python function for inference via mlflow.pyfunc.load_model().
You can also use the mlflow.sentence_transformers.load_model() function to load a saved or logged MLflow
Model with the sentence_transformers flavor as a native sentence-transformers model.
Example:
from sentence_transformers import SentenceTransformer
import mlflow
import mlflow.sentence_transformers
model = SentenceTransformer("all-MiniLM-L6-v2")
example_sentences = ["This is a sentence.", "This is another sentence."]
# Define the signature
signature = mlflow.models.infer_signature(
model_input=example_sentences,
model_output=model.encode(example_sentences),
)
# Log the model using mlflow
with mlflow.start_run():
logged_model = mlflow.sentence_transformers.log_model(
model=model,
artifact_path="sbert_model",
signature=signature,
input_example=example_sentences,
)
# Load option 1: mlflow.pyfunc.load_model returns a PyFuncModel
loaded_model = mlflow.pyfunc.load_model(logged_model.model_uri)
embeddings1 = loaded_model.predict(["hello world", "i am mlflow"])
# Load option 2: mlflow.sentence_transformers.load_model returns a SentenceTransformer
loaded_model = mlflow.sentence_transformers.load_model(logged_model.model_uri)
embeddings2 = loaded_model.encode(["hello world", "i am mlflow"])
print(embeddings1)
"""
>> [[-3.44772562e-02 3.10232025e-02 6.73496164e-03 2.61089969e-02
...
2.37922110e-02 -2.28897743e-02 3.89375277e-02 3.02067865e-02]
[ 4.81191138e-03 -9.33756605e-02 6.95968643e-02 8.09735525e-03
...
6.57437667e-02 -2.72239652e-02 4.02687863e-02 -1.05599344e-01]]
"""
Promptflow (promptflow) (Experimental)
Attention
The promptflow flavor is in active development and is marked as Experimental. Public APIs may change and new
features are subject to be added as additional functionality is brought to the flavor.
The promptflow model flavor is capable of packaging your flow in MLflow format via the mlflow.promptflow.save_model()
and mlflow.promptflow.log_model() functions. Currently, a flow.dag.yaml file is required to be
present in the flow’s directory. These functions also add the python_function flavor to the MLflow Models,
allowing the models to be interpreted as generic Python functions for inference via
mlflow.pyfunc.load_model(). You can also use the mlflow.promptflow.load_model()
method to load MLflow Models with the promptflow model flavor in native promptflow format.
Please note that the signature in MLmodel file will NOT BE automatically inferred from the flow itself.
To save model with the signature, you can either pass the input_example or specify the input signature manually.
Example:
Reach the flow source at example from the MLflow GitHub Repository.
import os
from pathlib import Path
from promptflow import load_flow
import mlflow
assert "OPENAI_API_KEY" in os.environ, "Please set the OPENAI_API_KEY environment variable."
# The example flow will write a simple code snippet that displays the greeting message with specific language.
flow_folder = Path(__file__).parent / "basic"
flow = load_flow(flow_folder)
with mlflow.start_run():
logged_model = mlflow.promptflow.log_model(flow, artifact_path="promptflow_model")
loaded_model = mlflow.pyfunc.load_model(logged_model.model_uri)
print(loaded_model.predict({"text": "Python Hello World!"}))
Model Evaluation
After building and training your MLflow Model, you can use the mlflow.evaluate() API to
evaluate its performance on one or more datasets of your choosing. mlflow.evaluate()
currently supports evaluation of MLflow Models with the
python_function (pyfunc) model flavor for classification, regression, and numerous language modeling tasks (see Evaluating with LLMs), computing a variety of
task-specific performance metrics, model performance plots, and
model explanations. Evaluation results are logged to MLflow Tracking.
The following example from the MLflow GitHub Repository
uses mlflow.evaluate() to evaluate the performance of a classifier
on the UCI Adult Data Set, logging a
comprehensive collection of MLflow Metrics and Artifacts that provide insight into model performance
and behavior:
import xgboost
import shap
import mlflow
from mlflow.models import infer_signature
from sklearn.model_selection import train_test_split
# Load the UCI Adult Dataset
X, y = shap.datasets.adult()
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
# Fit an XGBoost binary classifier on the training data split
model = xgboost.XGBClassifier().fit(X_train, y_train)
# Create a model signature
signature = infer_signature(X_test, model.predict(X_test))
# Build the Evaluation Dataset from the test set
eval_data = X_test
eval_data["label"] = y_test
with mlflow.start_run() as run:
# Log the baseline model to MLflow
mlflow.sklearn.log_model(model, "model", signature=signature)
model_uri = mlflow.get_artifact_uri("model")
# Evaluate the logged model
result = mlflow.evaluate(
model_uri,
eval_data,
targets="label",
model_type="classifier",
evaluators=["default"],
)
Evaluating with LLMs
As of MLflow 2.4.0, mlflow.evaluate() has built-in support for a variety of tasks with
LLMs, including text summarization, text classification, question answering, and text generation.
The following example uses mlflow.evaluate() to evaluate a model that answers
questions about MLflow (note that you must have the OPENAI_API_TOKEN environment variable set
in your current system environment in order to run the example):
import os
import pandas as pd
import mlflow
import openai
# Create a question answering model using prompt engineering with OpenAI. Log the
# prompt and the model to MLflow Tracking
mlflow.start_run()
system_prompt = (
"Your job is to answer questions about MLflow. When you are asked a question about MLflow,"
" respond to it. Make sure to include code examples. If the question is not related to"
" MLflow, refuse to answer and say that the question is unrelated."
)
mlflow.log_param("system_prompt", system_prompt)
logged_model = mlflow.openai.log_model(
model="gpt-4o-mini",
task=openai.chat.completions,
artifact_path="model",
messages=[
{"role": "system", "content": system_prompt},
{"role": "user", "content": "{question}"},
],
)
# Evaluate the model on some example questions
questions = pd.DataFrame(
{
"question": [
"How do you create a run with MLflow?",
"How do you log a model with MLflow?",
"What is the capital of France?",
]
}
)
mlflow.evaluate(
model=logged_model.model_uri,
model_type="question-answering",
data=questions,
)
# Load and inspect the evaluation results
results: pd.DataFrame = mlflow.load_table(
"eval_results_table.json", extra_columns=["run_id", "params.system_prompt"]
)
print("Evaluation results:")
print(results)
MLflow also provides an Artifact View UI for comparing inputs and outputs across multiple models built with LLMs. For example, after evaluating multiple prompts for question answering (see the MLflow OpenAI question answering full example), you can navigate to the Artifact View to view the questions and compare the answers for each model:
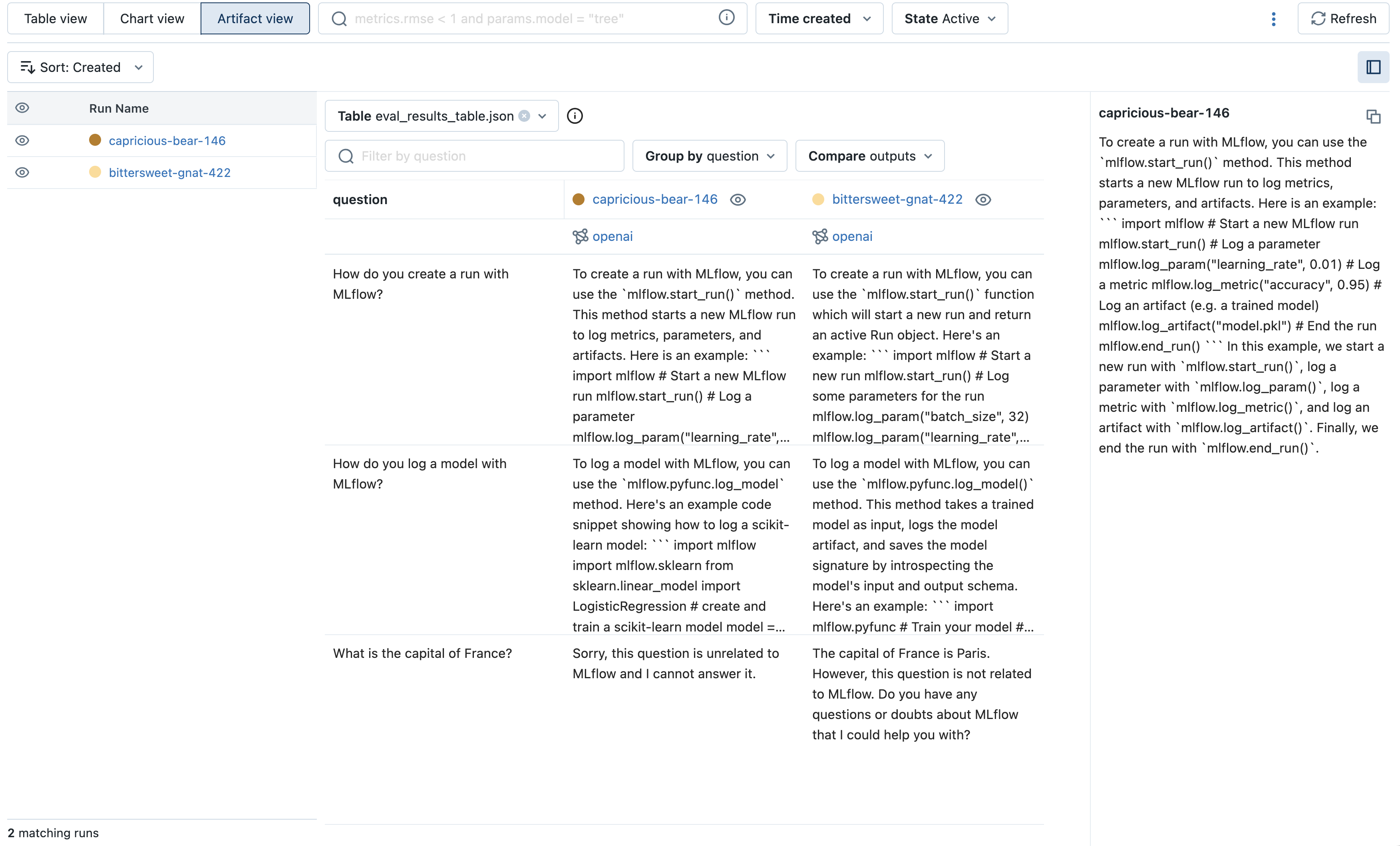
For additional examples demonstrating the use of mlflow.evaluate() with LLMs, check out the
MLflow LLMs example repository.
Evaluating with Extra Metrics
If the default set of metrics is insufficient, you can supply extra_metrics and custom_artifacts
to mlflow.evaluate() to produce extra metrics and artifacts for the model(s) that you’re evaluating.
To define an extra metric, you should define an eval_fn function that takes in predictions and targets as arguments
and outputs a MetricValue object. predictions and targets are pandas.Series
objects. If predictions or targets specified in mlflow.evaluate() is either numpy.array or List,
they will be converted to pandas.Series.
To use values from other metrics to compute your custom metric, include the name of the metric as an argument to eval_fn.
This argument will contain a MetricValue object which contains the values calculated from the specified metric and can be used to compute your custom metric.
{
"accuracy_score": MetricValue(
scores=None, justifications=None, aggregate_results={"accuracy_score": 1.0}
)
}
The MetricValue class has three attributes:
scores: a list that contains per-row metrics.aggregate_results: a dictionary that maps the aggregation method names to the corresponding aggregated values. This is intended to be used to aggregatescores.justifications: a list that contains per-row justifications of the values inscores. This is optional, and is usually used with genai metrics.
The code block below demonstrates how to define a custom metric evaluation function:
from mlflow.metrics import MetricValue
def my_metric_eval_fn(predictions, targets):
scores = np.abs(predictions - targets)
return MetricValue(
scores=list(scores),
aggregate_results={
"mean": np.mean(scores),
"variance": np.var(scores),
"median": np.median(scores),
},
)
Once you have defined an eval_fn, you then use make_metric() to wrap this eval_fn function into a metric.
In addition to eval_fn, make_metric() requires an additional parameter , greater_is_better, for optimization purposes. This parameter
indicates whether this is a metric we want to maximize or minimize.
from mlflow.metrics import make_metric
mymetric = make_metric(eval_fn=my_metric_eval_fn, greater_is_better=False)
The extra metric allows you to either evaluate a model directly, or to evaluate an output dataframe.
To evaluate the model directly, you will have to provide mlflow.evaluate() either a pyfunc model
instance, a URI referring to a pyfunc model, or a callable function that takes in the data as input
and outputs the predictions.
def model(x):
return x["inputs"]
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"inputs": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
mlflow.evaluate(model, eval_dataset, targets="targets", extra_metrics=[mymetric])
- To directly evaluate an output dataframe, you can omit the
modelparameter. However, you will need to set the
predictionsparameter inmlflow.evaluate()in order to evaluate an inference output dataframe.
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"predictions": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
result = mlflow.evaluate(
data=eval_dataset,
predictions="predictions",
targets="targets",
extra_metrics=[mymetric],
)
When your model has multiple outputs, the model must return a pandas DataFrame with multiple columns. You must
specify one column among the model output columns as the predictions column using the predictions parameter,
and other output columns of the model will be accessible from the eval_fn based on their column names. For example, if
your model has two outputs retrieved_context and answer, you can specify answer as the predictions
column, and retrieved_context column will be accessible as the context parameter from eval_fn via col_mapping:
def eval_fn(predictions, targets, context):
scores = (predictions == targets) + context
return MetricValue(
scores=list(scores),
aggregate_results={"mean": np.mean(scores), "sum": np.sum(scores)},
)
mymetric = make_metric(eval_fn=eval_fn, greater_is_better=False, name="mymetric")
def model(x):
return pd.DataFrame({"retrieved_context": x["inputs"] + 1, "answer": x["inputs"]})
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"inputs": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
config = {"col_mapping": {"context": "retrieved_context"}}
result = mlflow.evaluate(
model,
eval_dataset,
predictions="answer",
targets="targets",
extra_metrics=[mymetric],
evaluator_config=config,
)
However, you can also avoid using col_mapping if the parameter of eval_fn is the same as the output column name of the model.
def eval_fn(predictions, targets, retrieved_context):
scores = (predictions == targets) + retrieved_context
return MetricValue(
scores=list(scores),
aggregate_results={"mean": np.mean(scores), "sum": np.sum(scores)},
)
mymetric = make_metric(eval_fn=eval_fn, greater_is_better=False, name="mymetric")
def model(x):
return pd.DataFrame({"retrieved_context": x["inputs"] + 1, "answer": x["inputs"]})
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"inputs": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
result = mlflow.evaluate(
model,
eval_dataset,
predictions="answer",
targets="targets",
extra_metrics=[mymetric],
)
col_mapping also allows you to pass additional parameters to the extra metric function, in this case passing a value k.
def eval_fn(predictions, targets, k):
scores = k * (predictions == targets)
return MetricValue(scores=list(scores), aggregate_results={"mean": np.mean(scores)})
weighted_mymetric = make_metric(eval_fn=eval_fn, greater_is_better=False)
def model(x):
return x["inputs"]
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"inputs": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
config = {"col_mapping": {"k": 5}}
mlflow.evaluate(
model,
eval_dataset,
targets="targets",
extra_metrics=[weighted_mymetric],
evaluator_config=config,
)
You can also add the name of other metrics as an argument to the extra metric function, which will pass in the MetricValue calculated for that metric.
def eval_fn(predictions, targets, retrieved_context):
scores = (predictions == targets) + retrieved_context
return MetricValue(
scores=list(scores),
aggregate_results={"mean": np.mean(scores), "sum": np.sum(scores)},
)
mymetric = make_metric(eval_fn=eval_fn, greater_is_better=False, name="mymetric")
def eval_fn_2(predictions, targets, mymetric):
scores = ["true" if score else "false" for score in mymetric.scores]
return MetricValue(
scores=list(scores),
)
mymetric2 = make_metric(eval_fn=eval_fn_2, greater_is_better=False, name="mymetric2")
def model(x):
return pd.DataFrame({"retrieved_context": x["inputs"] + 1, "answer": x["inputs"]})
eval_dataset = pd.DataFrame(
{
"targets": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
"inputs": [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0],
}
)
result = mlflow.evaluate(
model,
eval_dataset,
predictions="answer",
targets="targets",
extra_metrics=[mymetric, mymetric2],
)
The following short example from the MLflow GitHub Repository
uses mlflow.evaluate() with an extra metric function to evaluate the performance of a regressor on the
California Housing Dataset.
import os
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import fetch_california_housing
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
import mlflow
from mlflow.models import infer_signature, make_metric
# loading the California housing dataset
cali_housing = fetch_california_housing(as_frame=True)
# split the dataset into train and test partitions
X_train, X_test, y_train, y_test = train_test_split(
cali_housing.data, cali_housing.target, test_size=0.2, random_state=123
)
# train the model
lin_reg = LinearRegression().fit(X_train, y_train)
# Infer model signature
predictions = lin_reg.predict(X_train)
signature = infer_signature(X_train, predictions)
# creating the evaluation dataframe
eval_data = X_test.copy()
eval_data["target"] = y_test
def squared_diff_plus_one(eval_df, _builtin_metrics):
"""
This example custom metric function creates a metric based on the ``prediction`` and
``target`` columns in ``eval_df`.
"""
return np.sum(np.abs(eval_df["prediction"] - eval_df["target"] + 1) ** 2)
def sum_on_target_divided_by_two(_eval_df, builtin_metrics):
"""
This example custom metric function creates a metric derived from existing metrics in
``builtin_metrics``.
"""
return builtin_metrics["sum_on_target"] / 2
def prediction_target_scatter(eval_df, _builtin_metrics, artifacts_dir):
"""
This example custom artifact generates and saves a scatter plot to ``artifacts_dir`` that
visualizes the relationship between the predictions and targets for the given model to a
file as an image artifact.
"""
plt.scatter(eval_df["prediction"], eval_df["target"])
plt.xlabel("Targets")
plt.ylabel("Predictions")
plt.title("Targets vs. Predictions")
plot_path = os.path.join(artifacts_dir, "example_scatter_plot.png")
plt.savefig(plot_path)
return {"example_scatter_plot_artifact": plot_path}
with mlflow.start_run() as run:
mlflow.sklearn.log_model(lin_reg, "model", signature=signature)
model_uri = mlflow.get_artifact_uri("model")
result = mlflow.evaluate(
model=model_uri,
data=eval_data,
targets="target",
model_type="regressor",
evaluators=["default"],
extra_metrics=[
make_metric(
eval_fn=squared_diff_plus_one,
greater_is_better=False,
),
make_metric(
eval_fn=sum_on_target_divided_by_two,
greater_is_better=True,
),
],
custom_artifacts=[prediction_target_scatter],
)
print(f"metrics:\n{result.metrics}")
print(f"artifacts:\n{result.artifacts}")
For a more comprehensive extra metrics usage example, refer to this example from the MLflow GitHub Repository.
Evaluating with a Function
As of MLflow 2.8.0, mlflow.evaluate() supports evaluating a python function without requiring
logging the model to MLflow. This is useful when you don’t want to log the model and just want to evaluate
it. The requirements for the function’s input and output are the same as the requirements for a model’s input and
output.
The following example uses mlflow.evaluate() to evaluate a function:
import shap
import xgboost
from sklearn.model_selection import train_test_split
import mlflow
# Load the UCI Adult Dataset
X, y = shap.datasets.adult()
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
# Fit an XGBoost binary classifier on the training data split
model = xgboost.XGBClassifier().fit(X_train, y_train)
# Build the Evaluation Dataset from the test set
eval_data = X_test
eval_data["label"] = y_test
# Define a function that calls the model's predict method
def fn(X):
return model.predict(X)
with mlflow.start_run() as run:
# Evaluate the function without logging the model
result = mlflow.evaluate(
fn,
eval_data,
targets="label",
model_type="classifier",
evaluators=["default"],
)
print(f"metrics:\n{result.metrics}")
print(f"artifacts:\n{result.artifacts}")
Evaluating with a Static Dataset
As of MLflow 2.8.0, mlflow.evaluate() supports evaluating a static dataset without specifying a model.
This is useful when you save the model output to a column in a Pandas DataFrame or an MLflow PandasDataset, and
want to evaluate the static dataset without re-running the model.
If you are using a Pandas DataFrame, you must specify the column name that contains the model output using the
top-level predictions parameter in mlflow.evaluate():
# Assume that the model output is saved to the pandas_df["model_output"] column
mlflow.evaluate(data=pandas_df, predictions="model_output", ...)
If you are using an MLflow PandasDataset, you must specify the column name that contains the model output using
the predictions parameter in mlflow.data.from_pandas(), and specify None for the
predictions parameter in mlflow.evaluate():
# Assume that the model output is saved to the pandas_df["model_output"] column
dataset = mlflow.data.from_pandas(pandas_df, predictions="model_output")
mlflow.evaluate(data=pandas_df, predictions=None, ...)
When your model has multiple outputs, you must specify one column among the model output columns as the predictions
column. The other output columns of the model will be treated as “input” columns. For example, if your model
has two outputs named retrieved_context and answer, you can specify answer as the predictions column. The
retrieved_context column will be treated as an “input” column when calculating the metrics.
The following example uses mlflow.evaluate() to evaluate a static dataset:
import shap
import xgboost
from sklearn.model_selection import train_test_split
import mlflow
# Load the UCI Adult Dataset
X, y = shap.datasets.adult()
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
# Fit an XGBoost binary classifier on the training data split
model = xgboost.XGBClassifier().fit(X_train, y_train)
# Build the Evaluation Dataset from the test set
y_test_pred = model.predict(X=X_test)
eval_data = X_test
eval_data["label"] = y_test
eval_data["predictions"] = y_test_pred
with mlflow.start_run() as run:
# Evaluate the static dataset without providing a model
result = mlflow.evaluate(
data=eval_data,
targets="label",
predictions="predictions",
model_type="classifier",
)
print(f"metrics:\n{result.metrics}")
print(f"artifacts:\n{result.artifacts}")
Performing Model Validation
Attention
MLflow 2.18.0 has moved the model validation functionality from the mlflow.evaluate() API to a dedicated mlflow.validate_evaluation_results() API. The relevant parameters, such as baseline_model, are deprecated and will be removed from the older API in future versions.
With the mlflow.validate_evaluation_results() API, you can validate metrics generated during model evaluation to assess the quality of your model against a baseline. To achieve this, first evaluate both the candidate and baseline models using mlflow.evaluate() (or load persisted evaluation results from local storage). Then, pass the results along with a validation_thresholds dictionary that maps metric names to mlflow.models.MetricThreshold objects. If your model fails to meet the specified thresholds, mlflow.validate_evaluation_results() will raise a ModelValidationFailedException with details about the validation failure.
More information on model evaluation behavior and outputs can be found in the mlflow.evaluate() API documentation.
Validating a Model Agasint a Baseline Model
Below is an example of how to validate a candidate model’s performance against a baseline using predefined thresholds.
import xgboost
import shap
from sklearn.model_selection import train_test_split
from sklearn.dummy import DummyClassifier
import mlflow
from mlflow.models import MetricThreshold
# load UCI Adult Data Set; segment it into training and test sets
X, y = shap.datasets.adult()
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
# construct an evaluation dataset from the test set
eval_data = X_test
eval_data["label"] = y_test
# Log and evaluate the candidate model
candidate_model = xgboost.XGBClassifier().fit(X_train, y_train)
with mlflow.start_run(run_name="candidate") as run:
candidate_model_uri = mlflow.sklearn.log_model(
candidate_model, "candidate_model", signature=signature
).model_uri
candidate_result = mlflow.evaluate(
candidate_model_uri,
eval_data,
targets="label",
model_type="classifier",
)
# Log and evaluate the baseline model
baseline_model = DummyClassifier(strategy="uniform").fit(X_train, y_train)
with mlflow.start_run(run_name="baseline") as run:
baseline_model_uri = mlflow.sklearn.log_model(
baseline_model, "baseline_model", signature=signature
).model_uri
baseline_result = mlflow.evaluate(
baseline_model_uri,
eval_data,
targets="label",
model_type="classifier",
)
# Define criteria for model to be validated against
thresholds = {
"accuracy_score": MetricThreshold(
threshold=0.8, # accuracy should be >=0.8
min_absolute_change=0.05, # accuracy should be at least 0.05 greater than baseline model accuracy
min_relative_change=0.05, # accuracy should be at least 5 percent greater than baseline model accuracy
greater_is_better=True,
),
}
# Validate the candidate model agaisnt baseline
mlflow.validate_evaluation_results(
candidate_result=candidate_result,
baseline_result=baseline_result,
validation_thresholds=thresholds,
)
Refer to mlflow.models.MetricThreshold to see details on how the thresholds are specified
and checked.
The logged output within the MLflow UI for the comprehensive example is shown below.
Note
Model validation results are not included in the active MLflow run.
Validating a Model Against Static Thresholds
The mlflow.validate_evaluation_results() API can also be used to validate a candidate model against static thresholds, instead of comparing it to a baseline model, by passing None to the baseline_result parameter.
import mlflow
from mlflow.models import MetricThreshold
thresholds = {
"accuracy_score": MetricThreshold(
threshold=0.8, # accuracy should be >=0.8
greater_is_better=True,
),
}
# Validate the candidate model agaisnt static threshold
mlflow.validate_evaluation_results(
candidate_result=candidate_result,
baseline_result=None,
validation_thresholds=thresholds,
)
Reusing Baseline Result For Multiple Validations
The mlflow.models.EvaluationResult object returned by mlflow.evaluate() can be saved to and loaded from local storage. This feature allows you to reuse the same baseline result across different candidate models, which is particularly useful for automating model quality monitoring.
import mlflow
from mlflow.models.evaluation import EvaluationResult
baseline_result = mlflow.evaluate(
baseline_model_uri,
eval_data,
targets="label",
model_type="classifier",
)
baseline_result.save("RESULT_PATH")
# Load the evaluation result for validation
baseline_result = EvaluationResult.load("RESULT_PATH")
Note
There are plugins that support in-depth model validation with features that are not supported directly in MLflow. To learn more, see:
Note
Differences in the computation of Area under Curve Precision Recall score (metric name
precision_recall_auc) between multi and binary classifiers:
Multiclass classifier models, when evaluated, utilize the standard scoring metric from sklearn:
sklearn.metrics.roc_auc_score to calculate the area under the precision recall curve. This
algorithm performs a linear interpolation calculation utilizing the trapezoidal rule to estimate
the area under the precision recall curve. It is well-suited for use in evaluating multi-class
classification models to provide a single numeric value of the quality of fit.
Binary classifier models, on the other hand, use the sklearn.metrics.average_precision_score to
avoid the shortcomings of the roc_auc_score implementation when applied to heavily
imbalanced classes in binary classification. Usage of the roc_auc_score for imbalanced
datasets can give a misleading result (optimistically better than the model’s actual ability
to accurately predict the minority class membership).
For additional information on the topic of why different algorithms are employed for this, as
well as links to the papers that informed the implementation of these metrics within the
sklearn.metrics module, refer to
the documentation.
For simplicity purposes, both methodologies evaluation metric results (whether for multi-class
or binary classification) are unified in the single metric: precision_recall_auc.
Model Validation with Giskard’s plugin
To extend the validation capabilities of MLflow and anticipate issues before they go to production, a plugin has been built by Giskard allowing users to:
scan a model in order to detect hidden vulnerabilities such as Performance bias, Unrobustness, Overconfidence, Underconfidence, Ethical bias, Data leakage, Stochasticity, Spurious correlation, and others
explore samples in the data that highlight the vulnerabilities found
log the vulnerabilities as well-defined and quantified metrics
compare the metrics across different models
See the following plugin example notebooks for a demo:
For more information on the plugin, see the giskard-mlflow docs.
Model Validation with Trubrics’ plugin
To extend the validation capabilities of MLflow, a plugin has been built by Trubrics allowing users:
to use a large number of out-of-the-box validations
to validate a run with any custom python functions
to view all validation results in a .json file, for diagnosis of why an MLflow run could have failed
See the plugin example notebook for a demo.
For more information on the plugin, see the trubrics-mlflow docs.
Model Customization
While MLflow’s built-in model persistence utilities are convenient for packaging models from various popular ML libraries in MLflow Model format, they do not cover every use case. For example, you may want to use a model from an ML library that is not explicitly supported by MLflow’s built-in flavors. Alternatively, you may want to package custom inference code and data to create an MLflow Model. Fortunately, MLflow provides two solutions that can be used to accomplish these tasks: Custom Python Models and Custom Flavors.
In this section:
Custom Python Models
The mlflow.pyfunc module provides save_model() and
log_model() utilities for creating MLflow Models with the
python_function flavor that contain user-specified code and artifact (file) dependencies.
These artifact dependencies may include serialized models produced by any Python ML library.
Because these custom models contain the python_function flavor, they can be deployed
to any of MLflow’s supported production environments, such as SageMaker, AzureML, or local
REST endpoints.
The following examples demonstrate how you can use the mlflow.pyfunc module to create
custom Python models. For additional information about model customization with MLflow’s
python_function utilities, see the
python_function custom models documentation.
Example: Creating a custom “add n” model
This example defines a class for a custom model that adds a specified numeric value, n, to all
columns of a Pandas DataFrame input. Then, it uses the mlflow.pyfunc APIs to save an
instance of this model with n = 5 in MLflow Model format. Finally, it loads the model in
python_function format and uses it to evaluate a sample input.
import mlflow.pyfunc
# Define the model class
class AddN(mlflow.pyfunc.PythonModel):
def __init__(self, n):
self.n = n
def predict(self, context, model_input, params=None):
return model_input.apply(lambda column: column + self.n)
# Construct and save the model
model_path = "add_n_model"
add5_model = AddN(n=5)
mlflow.pyfunc.save_model(path=model_path, python_model=add5_model)
# Load the model in `python_function` format
loaded_model = mlflow.pyfunc.load_model(model_path)
# Evaluate the model
import pandas as pd
model_input = pd.DataFrame([range(10)])
model_output = loaded_model.predict(model_input)
assert model_output.equals(pd.DataFrame([range(5, 15)]))
Example: Saving an XGBoost model in MLflow format
This example begins by training and saving a gradient boosted tree model using the XGBoost
library. Next, it defines a wrapper class around the XGBoost model that conforms to MLflow’s
python_function inference API. Then, it uses the wrapper class and
the saved XGBoost model to construct an MLflow Model that performs inference using the gradient
boosted tree. Finally, it loads the MLflow Model in python_function format and uses it to
evaluate test data.
# Load training and test datasets
from sys import version_info
import xgboost as xgb
from sklearn import datasets
from sklearn.model_selection import train_test_split
PYTHON_VERSION = f"{version_info.major}.{version_info.minor}.{version_info.micro}"
iris = datasets.load_iris()
x = iris.data[:, 2:]
y = iris.target
x_train, x_test, y_train, _ = train_test_split(x, y, test_size=0.2, random_state=42)
dtrain = xgb.DMatrix(x_train, label=y_train)
# Train and save an XGBoost model
xgb_model = xgb.train(params={"max_depth": 10}, dtrain=dtrain, num_boost_round=10)
xgb_model_path = "xgb_model.pth"
xgb_model.save_model(xgb_model_path)
# Create an `artifacts` dictionary that assigns a unique name to the saved XGBoost model file.
# This dictionary will be passed to `mlflow.pyfunc.save_model`, which will copy the model file
# into the new MLflow Model's directory.
artifacts = {"xgb_model": xgb_model_path}
# Define the model class
import mlflow.pyfunc
class XGBWrapper(mlflow.pyfunc.PythonModel):
def load_context(self, context):
import xgboost as xgb
self.xgb_model = xgb.Booster()
self.xgb_model.load_model(context.artifacts["xgb_model"])
def predict(self, context, model_input, params=None):
input_matrix = xgb.DMatrix(model_input.values)
return self.xgb_model.predict(input_matrix)
# Create a Conda environment for the new MLflow Model that contains all necessary dependencies.
import cloudpickle
conda_env = {
"channels": ["defaults"],
"dependencies": [
f"python={PYTHON_VERSION}",
"pip",
{
"pip": [
f"mlflow=={mlflow.__version__}",
f"xgboost=={xgb.__version__}",
f"cloudpickle=={cloudpickle.__version__}",
],
},
],
"name": "xgb_env",
}
# Save the MLflow Model
mlflow_pyfunc_model_path = "xgb_mlflow_pyfunc"
mlflow.pyfunc.save_model(
path=mlflow_pyfunc_model_path,
python_model=XGBWrapper(),
artifacts=artifacts,
conda_env=conda_env,
)
# Load the model in `python_function` format
loaded_model = mlflow.pyfunc.load_model(mlflow_pyfunc_model_path)
# Evaluate the model
import pandas as pd
test_predictions = loaded_model.predict(pd.DataFrame(x_test))
print(test_predictions)
Example: Logging a transformers model with hf:/ schema to avoid copying large files
This example shows how to use a special schema hf:/ to log a transformers model from huggingface
hub directly. This is useful when the model is too large and especially when you want to serve the
model directly, but it doesn’t save extra space if you want to download and test the model locally.
import mlflow
from mlflow.models import infer_signature
import numpy as np
import transformers
# Define a custom PythonModel
class QAModel(mlflow.pyfunc.PythonModel):
def load_context(self, context):
"""
This method initializes the tokenizer and language model
using the specified snapshot location from model context.
"""
snapshot_location = context.artifacts["bert-tiny-model"]
# Initialize tokenizer and language model
tokenizer = transformers.AutoTokenizer.from_pretrained(snapshot_location)
model = transformers.BertForQuestionAnswering.from_pretrained(snapshot_location)
self.pipeline = transformers.pipeline(
task="question-answering", model=model, tokenizer=tokenizer
)
def predict(self, context, model_input, params=None):
question = model_input["question"][0]
if isinstance(question, np.ndarray):
question = question.item()
ctx = model_input["context"][0]
if isinstance(ctx, np.ndarray):
ctx = ctx.item()
return self.pipeline(question=question, context=ctx)
# Log the model
data = {"question": "Who's house?", "context": "The house is owned by Run."}
pyfunc_artifact_path = "question_answering_model"
with mlflow.start_run() as run:
model_info = mlflow.pyfunc.log_model(
artifact_path=pyfunc_artifact_path,
python_model=QAModel(),
artifacts={"bert-tiny-model": "hf:/prajjwal1/bert-tiny"},
input_example=data,
signature=infer_signature(data, ["Run"]),
extra_pip_requirements=["torch", "accelerate", "transformers", "numpy"],
)
Custom Flavors
You can also create custom MLflow Models by writing a custom flavor.
As discussed in the Model API and Storage Format sections, an MLflow Model
is defined by a directory of files that contains an MLmodel configuration file. This MLmodel
file describes various model attributes, including the flavors in which the model can be
interpreted. The MLmodel file contains an entry for each flavor name; each entry is
a YAML-formatted collection of flavor-specific attributes.
To create a new flavor to support a custom model, you define the set of flavor-specific attributes
to include in the MLmodel configuration file, as well as the code that can interpret the
contents of the model directory and the flavor’s attributes. A detailed example of constructing a
custom model flavor and its usage is shown below. New custom flavors not considered for official
inclusion into MLflow should be introduced as separate GitHub repositories with documentation
provided in the
Community Model Flavors
page.
Example: Creating a custom “sktime” flavor
This example illustrates the creation of a custom flavor for
sktime time series library. The library provides a unified
interface for multiple learning tasks including time series forecasting. While the custom flavor in
this example is specific in terms of the sktime inference API and model serialization format,
its interface design is similar to many of the existing built-in flavors. Particularly, the
interface for utilizing the custom model loaded as a python_function flavor for generating
predictions uses a single-row Pandas DataFrame configuration argument to expose the paramters
of the sktime inference API. The complete code for this example is included in the
flavor.py module of the
sktime example directory.
Let’s examine the custom flavor module in more detail. The first step is to import several modules
inluding sktime library, various MLflow utilities as well as the MLflow pyfunc module which
is required to add the pyfunc specification to the MLflow model configuration. Note also the
import of the flavor module itself. This will be passed to the
mlflow.models.Model.log() method to log the model as an artifact to the current MLflow
run.
import logging
import os
import pickle
import flavor
import mlflow
import numpy as np
import pandas as pd
import sktime
import yaml
from mlflow import pyfunc
from mlflow.exceptions import MlflowException
from mlflow.models import Model
from mlflow.models.model import MLMODEL_FILE_NAME
from mlflow.models.utils import _save_example
from mlflow.protos.databricks_pb2 import INTERNAL_ERROR, INVALID_PARAMETER_VALUE
from mlflow.tracking._model_registry import DEFAULT_AWAIT_MAX_SLEEP_SECONDS
from mlflow.tracking.artifact_utils import _download_artifact_from_uri
from mlflow.utils.environment import (
_CONDA_ENV_FILE_NAME,
_CONSTRAINTS_FILE_NAME,
_PYTHON_ENV_FILE_NAME,
_REQUIREMENTS_FILE_NAME,
_mlflow_conda_env,
_process_conda_env,
_process_pip_requirements,
_PythonEnv,
_validate_env_arguments,
)
from mlflow.utils.file_utils import write_to
from mlflow.utils.model_utils import (
_add_code_from_conf_to_system_path,
_get_flavor_configuration,
_validate_and_copy_code_paths,
_validate_and_prepare_target_save_path,
)
from mlflow.utils.requirements_utils import _get_pinned_requirement
from sktime.utils.multiindex import flatten_multiindex
_logger = logging.getLogger(__name__)
We continue by defining a set of important variables used throughout the code that follows.
The flavor name needs to be provided for every custom flavor and should reflect the name of the
library to be supported. It is saved as part of the flavor-specific attributes to the MLmodel
configuration file. This example also defines some sktime specific variables. For illustration
purposes, only a subset of the available predict methods to be exposed via the
_SktimeModelWrapper class is included when loading the model in its python_function flavor
(additional methods could be added in a similar fashion). Additionaly, the model serialization
formats, namely pickle (default) and cloudpickle, are defined. Note that both serialization
modules require using the same python environment (version) in whatever environment this model is
used for inference to ensure that the model will load with the appropriate version of
pickle (cloudpickle).
FLAVOR_NAME = "sktime"
SKTIME_PREDICT = "predict"
SKTIME_PREDICT_INTERVAL = "predict_interval"
SKTIME_PREDICT_QUANTILES = "predict_quantiles"
SKTIME_PREDICT_VAR = "predict_var"
SUPPORTED_SKTIME_PREDICT_METHODS = [
SKTIME_PREDICT,
SKTIME_PREDICT_INTERVAL,
SKTIME_PREDICT_QUANTILES,
SKTIME_PREDICT_VAR,
]
SERIALIZATION_FORMAT_PICKLE = "pickle"
SERIALIZATION_FORMAT_CLOUDPICKLE = "cloudpickle"
SUPPORTED_SERIALIZATION_FORMATS = [
SERIALIZATION_FORMAT_PICKLE,
SERIALIZATION_FORMAT_CLOUDPICKLE,
]
Similar to the MLflow built-in flavors, a custom flavor logs the model in MLflow format via the
save_model() and log_model() functions. In the save_model() function, the sktime
model is saved to a specified output directory. Additionally, save_model() leverages the
mlflow.models.Model.add_flavor() and mlflow.models.Model.save() methods to
produce the MLmodel configuration containing the sktime and the python_function flavor.
The resulting configuration has several flavor-specific attributes, such as the flavor name and
sktime_version, which denotes the version of the sktime library that was used to train the
model. An example of the output directoy for the custom sktime model is shown below:
# Directory written by flavor.save_model(model, "my_model")
my_model/
├── MLmodel
├── conda.yaml
├── model.pkl
├── python_env.yaml
└── requirements.txt
And its YAML-formatted MLmodel file describes the two flavors:
flavors:
python_function:
env:
conda: conda.yaml
virtualenv: python_env.yaml
loader_module: flavor
model_path: model.pkl
python_version: 3.8.15
sktime:
code: null
pickled_model: model.pkl
serialization_format: pickle
sktime_version: 0.16.0
The save_model() function also provides flexibility to add additional paramters which can be
added as flavor-specific attributes to the model configuration. In this example there is only one
flavor-specific parameter for specifying the model serialization format. All other paramters are
non-flavor specific (for a detailed description of these parameters take a look at
mlflow.sklearn.save_model).
Note: When creating your own custom flavor, be sure rename the sktime_model parameter in both the
save_model() and log_model() functions to reflect the name of your custom model flavor.
def save_model(
sktime_model,
path,
conda_env=None,
code_paths=None,
mlflow_model=None,
signature=None,
input_example=None,
pip_requirements=None,
extra_pip_requirements=None,
serialization_format=SERIALIZATION_FORMAT_PICKLE,
):
_validate_env_arguments(conda_env, pip_requirements, extra_pip_requirements)
if serialization_format not in SUPPORTED_SERIALIZATION_FORMATS:
raise MlflowException(
message=(
f"Unrecognized serialization format: {serialization_format}. "
"Please specify one of the following supported formats: "
"{SUPPORTED_SERIALIZATION_FORMATS}."
),
error_code=INVALID_PARAMETER_VALUE,
)
_validate_and_prepare_target_save_path(path)
code_dir_subpath = _validate_and_copy_code_paths(code_paths, path)
if mlflow_model is None:
mlflow_model = Model()
if signature is not None:
mlflow_model.signature = signature
if input_example is not None:
_save_example(mlflow_model, input_example, path)
model_data_subpath = "model.pkl"
model_data_path = os.path.join(path, model_data_subpath)
_save_model(
sktime_model, model_data_path, serialization_format=serialization_format
)
pyfunc.add_to_model(
mlflow_model,
loader_module="flavor",
model_path=model_data_subpath,
conda_env=_CONDA_ENV_FILE_NAME,
python_env=_PYTHON_ENV_FILE_NAME,
code=code_dir_subpath,
)
mlflow_model.add_flavor(
FLAVOR_NAME,
pickled_model=model_data_subpath,
sktime_version=sktime.__version__,
serialization_format=serialization_format,
code=code_dir_subpath,
)
mlflow_model.save(os.path.join(path, MLMODEL_FILE_NAME))
if conda_env is None:
if pip_requirements is None:
include_cloudpickle = (
serialization_format == SERIALIZATION_FORMAT_CLOUDPICKLE
)
default_reqs = get_default_pip_requirements(include_cloudpickle)
inferred_reqs = mlflow.models.infer_pip_requirements(
path, FLAVOR_NAME, fallback=default_reqs
)
default_reqs = sorted(set(inferred_reqs).union(default_reqs))
else:
default_reqs = None
conda_env, pip_requirements, pip_constraints = _process_pip_requirements(
default_reqs, pip_requirements, extra_pip_requirements
)
else:
conda_env, pip_requirements, pip_constraints = _process_conda_env(conda_env)
with open(os.path.join(path, _CONDA_ENV_FILE_NAME), "w") as f:
yaml.safe_dump(conda_env, stream=f, default_flow_style=False)
if pip_constraints:
write_to(os.path.join(path, _CONSTRAINTS_FILE_NAME), "\n".join(pip_constraints))
write_to(os.path.join(path, _REQUIREMENTS_FILE_NAME), "\n".join(pip_requirements))
_PythonEnv.current().to_yaml(os.path.join(path, _PYTHON_ENV_FILE_NAME))
def _save_model(model, path, serialization_format):
with open(path, "wb") as out:
if serialization_format == SERIALIZATION_FORMAT_PICKLE:
pickle.dump(model, out)
else:
import cloudpickle
cloudpickle.dump(model, out)
The save_model() function also writes the model dependencies to a requirements.txt and
conda.yaml file in the model output directory. For this purpose the set of pip dependecies
produced by this flavor need to be added to the get_default_pip_requirements() function. In this
example only the minimum required dependencies are provided. In practice, additional requirements needed for
preprocessing or post-processing steps could be included. Note that for any custom flavor, the
mlflow.models.infer_pip_requirements() method in the save_model() function will
return the default requirements defined in get_default_pip_requirements() as package imports are
only inferred for built-in flavors.
def get_default_pip_requirements(include_cloudpickle=False):
pip_deps = [_get_pinned_requirement("sktime")]
if include_cloudpickle:
pip_deps += [_get_pinned_requirement("cloudpickle")]
return pip_deps
def get_default_conda_env(include_cloudpickle=False):
return _mlflow_conda_env(
additional_pip_deps=get_default_pip_requirements(include_cloudpickle)
)
Next, we add the log_model() function. This function is little more than a wrapper around the
mlflow.models.Model.log() method to enable logging our custom model as an artifact to the
curren MLflow run. Any flavor-specific parameters (e.g. serialization_format) introduced in the
save_model() function also need to be added in the log_model() function. We also need to
pass the flavor module to the mlflow.models.Model.log() method which internally calls
the save_model() function from above to persist the model.
def log_model(
sktime_model,
artifact_path,
conda_env=None,
code_paths=None,
registered_model_name=None,
signature=None,
input_example=None,
await_registration_for=DEFAULT_AWAIT_MAX_SLEEP_SECONDS,
pip_requirements=None,
extra_pip_requirements=None,
serialization_format=SERIALIZATION_FORMAT_PICKLE,
**kwargs,
):
return Model.log(
artifact_path=artifact_path,
flavor=flavor,
registered_model_name=registered_model_name,
sktime_model=sktime_model,
conda_env=conda_env,
code_paths=code_paths,
signature=signature,
input_example=input_example,
await_registration_for=await_registration_for,
pip_requirements=pip_requirements,
extra_pip_requirements=extra_pip_requirements,
serialization_format=serialization_format,
**kwargs,
)
To interpret model directories produced by save_model(), the custom flavor must also define a
load_model() function. The load_model() function reads the MLmodel configuration from
the specified model directory and uses the configuration attributes to load and return the
sktime model from its serialized representation.
def load_model(model_uri, dst_path=None):
local_model_path = _download_artifact_from_uri(
artifact_uri=model_uri, output_path=dst_path
)
flavor_conf = _get_flavor_configuration(
model_path=local_model_path, flavor_name=FLAVOR_NAME
)
_add_code_from_conf_to_system_path(local_model_path, flavor_conf)
sktime_model_file_path = os.path.join(
local_model_path, flavor_conf["pickled_model"]
)
serialization_format = flavor_conf.get(
"serialization_format", SERIALIZATION_FORMAT_PICKLE
)
return _load_model(
path=sktime_model_file_path, serialization_format=serialization_format
)
def _load_model(path, serialization_format):
with open(path, "rb") as pickled_model:
if serialization_format == SERIALIZATION_FORMAT_PICKLE:
return pickle.load(pickled_model)
elif serialization_format == SERIALIZATION_FORMAT_CLOUDPICKLE:
import cloudpickle
return cloudpickle.load(pickled_model)
The _load_pyfunc() function will be called by the mlflow.pyfunc.load_model() method
to load the custom model flavor as a pyfunc type. The MLmodel flavor configuration is used to
pass any flavor-specific attributes to the _load_model() function (i.e., the path to the
python_function flavor in the model directory and the model serialization format).
def _load_pyfunc(path):
try:
sktime_flavor_conf = _get_flavor_configuration(
model_path=path, flavor_name=FLAVOR_NAME
)
serialization_format = sktime_flavor_conf.get(
"serialization_format", SERIALIZATION_FORMAT_PICKLE
)
except MlflowException:
_logger.warning(
"Could not find sktime flavor configuration during model "
"loading process. Assuming 'pickle' serialization format."
)
serialization_format = SERIALIZATION_FORMAT_PICKLE
pyfunc_flavor_conf = _get_flavor_configuration(
model_path=path, flavor_name=pyfunc.FLAVOR_NAME
)
path = os.path.join(path, pyfunc_flavor_conf["model_path"])
return _SktimeModelWrapper(
_load_model(path, serialization_format=serialization_format)
)
The final step is to create the model wrapper class defining the python_function flavor. The
design of the wrapper class determines how the flavor’s inference API is exposed when making
predictions using the python_function flavor. Just like the built-in flavors, the predict()
method of the sktime wrapper class accepts a single-row Pandas DataFrame configuration
argument. For an example of how to construct this configuration DataFrame refer to the usage example
in the next section. A detailed description of the supported paramaters and input formats is
provided in the
flavor.py module
docstrings.
class _SktimeModelWrapper:
def __init__(self, sktime_model):
self.sktime_model = sktime_model
def predict(self, dataframe, params=None) -> pd.DataFrame:
df_schema = dataframe.columns.values.tolist()
if len(dataframe) > 1:
raise MlflowException(
f"The provided prediction pd.DataFrame contains {len(dataframe)} rows. "
"Only 1 row should be supplied.",
error_code=INVALID_PARAMETER_VALUE,
)
# Convert the configuration dataframe into a dictionary to simplify the
# extraction of parameters passed to the sktime predcition methods.
attrs = dataframe.to_dict(orient="index").get(0)
predict_method = attrs.get("predict_method")
if not predict_method:
raise MlflowException(
f"The provided prediction configuration pd.DataFrame columns ({df_schema}) do not "
"contain the required column `predict_method` for specifying the prediction method.",
error_code=INVALID_PARAMETER_VALUE,
)
if predict_method not in SUPPORTED_SKTIME_PREDICT_METHODS:
raise MlflowException(
"Invalid `predict_method` value."
f"The supported prediction methods are {SUPPORTED_SKTIME_PREDICT_METHODS}",
error_code=INVALID_PARAMETER_VALUE,
)
# For inference parameters 'fh', 'X', 'coverage', 'alpha', and 'cov'
# the respective sktime default value is used if the value was not
# provided in the configuration dataframe.
fh = attrs.get("fh", None)
# Any model that is trained with exogenous regressor elements will need
# to provide `X` entries as a numpy ndarray to the predict method.
X = attrs.get("X", None)
# When the model is served via REST API the exogenous regressor must be
# provided as a list to the configuration DataFrame to be JSON serializable.
# Below we convert the list back to ndarray type as required by sktime
# predict methods.
if isinstance(X, list):
X = np.array(X)
# For illustration purposes only a subset of the available sktime prediction
# methods is exposed. Additional methods (e.g. predict_proba) could be added
# in a similar fashion.
if predict_method == SKTIME_PREDICT:
predictions = self.sktime_model.predict(fh=fh, X=X)
if predict_method == SKTIME_PREDICT_INTERVAL:
coverage = attrs.get("coverage", 0.9)
predictions = self.sktime_model.predict_interval(
fh=fh, X=X, coverage=coverage
)
if predict_method == SKTIME_PREDICT_QUANTILES:
alpha = attrs.get("alpha", None)
predictions = self.sktime_model.predict_quantiles(fh=fh, X=X, alpha=alpha)
if predict_method == SKTIME_PREDICT_VAR:
cov = attrs.get("cov", False)
predictions = self.sktime_model.predict_var(fh=fh, X=X, cov=cov)
# Methods predict_interval() and predict_quantiles() return a pandas
# MultiIndex column structure. As MLflow signature inference does not
# support MultiIndex column structure the columns must be flattened.
if predict_method in [SKTIME_PREDICT_INTERVAL, SKTIME_PREDICT_QUANTILES]:
predictions.columns = flatten_multiindex(predictions)
return predictions
Example: Using the custom “sktime” flavor
This example trains a sktime NaiveForecaster model using the Longley dataset for forecasting
with exogenous variables. It shows a custom model type implementation that logs the training
hyper-parameters, evaluation metrics and the trained model as an artifact. The single-row
configuration DataFrame for this example defines an interval forecast with nominal coverage values
[0.9,0.95], a future forecast horizon of four periods, and an exogenous regressor.
import json
import flavor
import pandas as pd
from sktime.datasets import load_longley
from sktime.forecasting.model_selection import temporal_train_test_split
from sktime.forecasting.naive import NaiveForecaster
from sktime.performance_metrics.forecasting import (
mean_absolute_error,
mean_absolute_percentage_error,
)
import mlflow
ARTIFACT_PATH = "model"
with mlflow.start_run() as run:
y, X = load_longley()
y_train, y_test, X_train, X_test = temporal_train_test_split(y, X)
forecaster = NaiveForecaster()
forecaster.fit(
y_train,
X=X_train,
)
# Extract parameters
parameters = forecaster.get_params()
# Evaluate model
y_pred = forecaster.predict(fh=[1, 2, 3, 4], X=X_test)
metrics = {
"mae": mean_absolute_error(y_test, y_pred),
"mape": mean_absolute_percentage_error(y_test, y_pred),
}
print(f"Parameters: \n{json.dumps(parameters, indent=2)}")
print(f"Metrics: \n{json.dumps(metrics, indent=2)}")
# Log parameters and metrics
mlflow.log_params(parameters)
mlflow.log_metrics(metrics)
# Log model using custom model flavor with pickle serialization (default).
flavor.log_model(
sktime_model=forecaster,
artifact_path=ARTIFACT_PATH,
serialization_format="pickle",
)
model_uri = mlflow.get_artifact_uri(ARTIFACT_PATH)
# Load model in native sktime flavor and pyfunc flavor
loaded_model = flavor.load_model(model_uri=model_uri)
loaded_pyfunc = flavor.pyfunc.load_model(model_uri=model_uri)
# Convert test data to 2D numpy array so it can be passed to pyfunc predict using
# a single-row Pandas DataFrame configuration argument
X_test_array = X_test.to_numpy()
# Create configuration DataFrame
predict_conf = pd.DataFrame(
[
{
"fh": [1, 2, 3, 4],
"predict_method": "predict_interval",
"coverage": [0.9, 0.95],
"X": X_test_array,
}
]
)
# Generate interval forecasts with native sktime flavor and pyfunc flavor
print(
f"\nNative sktime 'predict_interval':\n${loaded_model.predict_interval(fh=[1, 2, 3], X=X_test, coverage=[0.9, 0.95])}"
)
print(f"\nPyfunc 'predict_interval':\n${loaded_pyfunc.predict(predict_conf)}")
# Print the run id wich is used for serving the model to a local REST API endpoint
print(f"\nMLflow run id:\n{run.info.run_id}")
When opening the MLflow runs detail page the serialized model artifact will show up, such as:
To serve the model to a local REST API endpoint run the following MLflow CLI command substituting the run id printed during execution of the previous block (for more details refer to the Deploy MLflow models section):
mlflow models serve -m runs:/<run_id>/model --env-manager local --host 127.0.0.1
An example of requesting a prediction from the served model is shown below. The exogenous regressor
needs to be provided as a list to be JSON serializable. The wrapper instance will convert the list
back to numpy ndarray type as required by sktime inference API.
import pandas as pd
import requests
from sktime.datasets import load_longley
from sktime.forecasting.model_selection import temporal_train_test_split
y, X = load_longley()
y_train, y_test, X_train, X_test = temporal_train_test_split(y, X)
# Define local host and endpoint url
host = "127.0.0.1"
url = f"http://{host}:5000/invocations"
# Create configuration DataFrame
X_test_list = X_test.to_numpy().tolist()
predict_conf = pd.DataFrame(
[
{
"fh": [1, 2, 3, 4],
"predict_method": "predict_interval",
"coverage": [0.9, 0.95],
"X": X_test_list,
}
]
)
# Create dictionary with pandas DataFrame in the split orientation
json_data = {"dataframe_split": predict_conf.to_dict(orient="split")}
# Score model
response = requests.post(url, json=json_data)
print(f"\nPyfunc 'predict_interval':\n${response.json()}")
Validate Models before Deployment
After logging your model with MLflow Tracking, it is highly recommended to validate the model locally before deploying it to production.
The mlflow.models.predict() API provides a convenient way to test your model in a virtual environment, offering isolated execution and several advantages:
Model dependencies validation: The API helps ensure that the dependencies logged with the model are correct and sufficient by executing the model with an input example in a virtual environment. For more details, refer to Validating Environment for Prediction.
Input data validation: The API can be used to validate the input data interacts with the model as expected by simulating the same data processing during model serving. Ensure that the input data is a valid example that aligns with the pyfunc model’s predict function requirements.
Extra environment variables validation: By specifying the
extra_envsparameter, you can test whether additional environment variables are required for the model to run successfully. Note that all existing environment variables inos.environare automatically passed into the virtual environment.
import mlflow
class MyModel(mlflow.pyfunc.PythonModel):
def predict(self, context, model_input, params=None):
return model_input
with mlflow.start_run():
model_info = mlflow.pyfunc.log_model(
"model",
python_model=MyModel(),
input_example=["a", "b", "c"],
)
mlflow.models.predict(
model_uri=model_info.model_uri,
input_data=["a", "b", "c"],
pip_requirements_override=["..."],
extra_envs={"MY_ENV_VAR": "my_value"},
)
Environment managers
The mlflow.models.predict() API supports the following environment managers to create the virtual environment for prediction:
virtualenv: The default environment manager.
uv: An extremely fast environment manager written in Rust. This is an experimental feature since MLflow 2.20.0.
conda: uses conda to create environment.
local: uses the current environment to run the model. Note thatpip_requirements_overrideis not supported in this mode.
Tip
Starting from MLflow 2.20.0, uv is available, and it is extremely fast.
Run pip install uv to install uv, or refer to uv installation guidance for other installation methods.
Example of using uv to create a virtual environment for prediction:
import mlflow
mlflow.models.predict(
model_uri="runs:/<run_id>/<model_path>",
input_data="your_data",
env_manager="uv",
)
Built-In Deployment Tools
This information is moved to MLflow Deployment page.
Export a python_function model as an Apache Spark UDF
Note
If you are using a model that has a very long running inference latency (i.e., a
transformers model) that could take longer than the default timeout of 60 seconds,
you can utilize the extra_env argument when defining the spark_udf instance for your
MLflow model, specifying an override to the environment variable MLFLOW_SCORING_SERVER_REQUEST_TIMEOUT.
For further guidance, please see mlflow.pyfunc.spark_udf().
You can output a python_function model as an Apache Spark UDF, which can be uploaded to a
Spark cluster and used to score the model.
Example
from pyspark.sql.functions import struct
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
pyfunc_udf = mlflow.pyfunc.spark_udf(spark, "<path-to-model>")
df = spark_df.withColumn("prediction", pyfunc_udf(struct([...])))
If a model contains a signature, the UDF can be called without specifying column name arguments. In this case, the UDF will be called with column names from signature, so the evaluation dataframe’s column names must match the model signature’s column names.
Example
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
pyfunc_udf = mlflow.pyfunc.spark_udf(spark, "<path-to-model-with-signature>")
df = spark_df.withColumn("prediction", pyfunc_udf())
If a model contains a signature with tensor spec inputs, you will need to pass a column of array type as a corresponding UDF argument. The values in this column must be comprised of one-dimensional arrays. The UDF will reshape the array values to the required shape with ‘C’ order (i.e. read / write the elements using C-like index order) and cast the values as the required tensor spec type. For example, assuming a model requires input ‘a’ of shape (-1, 2, 3) and input ‘b’ of shape (-1, 4, 5). In order to perform inference on this data, we need to prepare a Spark DataFrame with column ‘a’ containing arrays of length 6 and column ‘b’ containing arrays of length 20. We can then invoke the UDF like following example code:
Example
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
# Assuming the model requires input 'a' of shape (-1, 2, 3) and input 'b' of shape (-1, 4, 5)
model_path = "<path-to-model-requiring-multidimensional-inputs>"
pyfunc_udf = mlflow.pyfunc.spark_udf(spark, model_path)
# The `spark_df` has column 'a' containing arrays of length 6 and
# column 'b' containing arrays of length 20
df = spark_df.withColumn("prediction", pyfunc_udf(struct("a", "b")))
The resulting UDF is based on Spark’s Pandas UDF and is currently limited to producing either a single
value, an array of values, or a struct containing multiple field values
of the same type per observation. By default, we return the first
numeric column as a double. You can control what result is returned by supplying result_type
argument. The following values are supported:
'int'or IntegerType: The leftmost integer that can fit inint32result is returned or an exception is raised if there are none.'long'or LongType: The leftmost long integer that can fit inint64result is returned or an exception is raised if there are none.ArrayType (IntegerType | LongType): Return all integer columns that can fit into the requested size.
'float'or FloatType: The leftmost numeric result cast tofloat32is returned or an exception is raised if there are no numeric columns.'double'or DoubleType: The leftmost numeric result cast todoubleis returned or an exception is raised if there are no numeric columns.ArrayType ( FloatType | DoubleType ): Return all numeric columns cast to the requested type. An exception is raised if there are no numeric columns.
'string'or StringType: Result is the leftmost column cast as string.ArrayType ( StringType ): Return all columns cast as string.
'bool'or'boolean'or BooleanType: The leftmost column cast toboolis returned or an exception is raised if the values cannot be coerced.'field1 FIELD1_TYPE, field2 FIELD2_TYPE, ...': A struct type containing multiple fields separated by comma, each field type must be one of types listed above.
Example
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
# Suppose the PyFunc model `predict` method returns a dict like:
# `{'prediction': 1-dim_array, 'probability': 2-dim_array}`
# You can supply result_type to be a struct type containing
# 2 fields 'prediction' and 'probability' like following.
pyfunc_udf = mlflow.pyfunc.spark_udf(
spark, "<path-to-model>", result_type="prediction float, probability: array<float>"
)
df = spark_df.withColumn("prediction", pyfunc_udf())
Example
from pyspark.sql.types import ArrayType, FloatType
from pyspark.sql.functions import struct
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
pyfunc_udf = mlflow.pyfunc.spark_udf(
spark, "path/to/model", result_type=ArrayType(FloatType())
)
# The prediction column will contain all the numeric columns returned by the model as floats
df = spark_df.withColumn("prediction", pyfunc_udf(struct("name", "age")))
If you want to use conda to restore the python environment that was used to train the model,
set the env_manager argument when calling mlflow.pyfunc.spark_udf().
Example
from pyspark.sql.types import ArrayType, FloatType
from pyspark.sql.functions import struct
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
pyfunc_udf = mlflow.pyfunc.spark_udf(
spark,
"path/to/model",
result_type=ArrayType(FloatType()),
env_manager="conda", # Use conda to restore the environment used in training
)
df = spark_df.withColumn("prediction", pyfunc_udf(struct("name", "age")))
If you want to call :py:func:`mlflow.pyfunc.spark_udf through Databricks connect in remote client, you need to build the model environment in Databricks runtime first.
Example
from mlflow.pyfunc import build_model_env
# Build the model env and save it as an archive file to the provided UC volume directory
# and print the saved model env archive file path (like '/Volumes/.../.../XXXXX.tar.gz')
print(build_model_env(model_uri, "/Volumes/..."))
# print the cluster id. Databricks Connect client needs to use the cluster id.
print(spark.conf.get("spark.databricks.clusterUsageTags.clusterId"))
Once you have pre-built the model environment, you can run :py:func:`mlflow.pyfunc.spark_udf with ‘prebuilt_model_env’ parameter through Databricks connect in remote client,
Example
from databricks.connect import DatabricksSession
spark = DatabricksSession.builder.remote(
host=os.environ["DATABRICKS_HOST"],
token=os.environ["DATABRICKS_TOKEN"],
cluster_id="<cluster id>", # get cluster id by spark.conf.get("spark.databricks.clusterUsageTags.clusterId")
).getOrCreate()
# The path generated by `build_model_env` in Databricks runtime.
model_env_uc_uri = "dbfs:/Volumes/.../.../XXXXX.tar.gz"
pyfunc_udf = mlflow.pyfunc.spark_udf(
spark, model_uri, prebuilt_env_uri=model_env_uc_uri
)
Deployment to Custom Targets
In addition to the built-in deployment tools, MLflow provides a pluggable mlflow.deployments Python API and mlflow deployments CLI for deploying models to custom targets and environments. To deploy to a custom target, you must first install an appropriate third-party Python plugin. See the list of known community-maintained plugins here.
Commands
The mlflow deployments CLI contains the following commands, which can also be invoked programmatically using the mlflow.deployments Python API:
Create: Deploy an MLflow model to a specified custom target
Delete: Delete a deployment
Update: Update an existing deployment, for example to deploy a new model version or change the deployment’s configuration (e.g. increase replica count)
List: List IDs of all deployments
Get: Print a detailed description of a particular deployment
Run Local: Deploy the model locally for testing
Help: Show the help string for the specified target
For more info, see:
mlflow deployments --help
mlflow deployments create --help
mlflow deployments delete --help
mlflow deployments update --help
mlflow deployments list --help
mlflow deployments get --help
mlflow deployments run-local --help
mlflow deployments help --help
Community Model Flavors
Go to the Community Model Flavors page to get an overview of other useful MLflow flavors, which are developed and maintained by the MLflow community.